Clostridium welchii specific detection primer and detection kit
A technology of Clostridium welchii disease and Clostridium welchii, which is applied to biochemical equipment and methods, measurement/inspection of microorganisms, microorganisms, etc., can solve the problems of low detection rate, cumbersome steps, long cycle, etc., and achieve PCR Appropriate reaction conditions, sensitive PCR detection, and objective result judgment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1 Primer Design and Screening
[0050] 1. Experimental steps
[0051] (1) According to the entire gene sequence encoding Clostridium welchii 16s rRNA, compare it with the 16s rRNA gene sequences of other bacteria, select specific genes as target genes for the diagnosis of Clostridioides welchii disease, and design specific primers according to their gene sequences, See Table 1.
[0052] Table 1 Clostridium welchii specific PCR primers
[0053]
[0054] (2) Extraction of fecal DNA: extraction was performed using a commercially available kit, and the specific steps were referred to the instructions of QIAamp Company.
[0055](3) The PCR reaction system is: Premix Ex Taq Version 2.0 (Loading dye Mix) 12.5 μL, forward primer and reverse primer 0.5 μL each, double distilled water 9.5 μL, template DNA 2 μL, the number of amplified samples n (n = number of samples + 3). Mix the above reaction solution in a centrifuge tube, mix well, and pack. Take the DNA of ea...
Embodiment 2
[0064] The optimization of embodiment 2 PCR reaction conditions
[0065] 1. Experimental steps
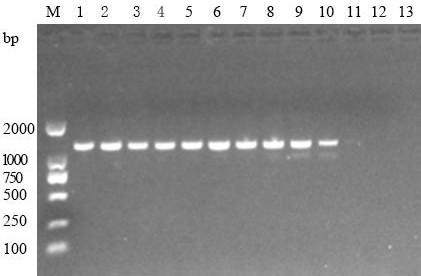
[0066] Use the primer TTF-2 / TTR-2 (the nucleotide series shown in SEQ ID NO: 1-2) screened in Example 1 to perform PCR amplification. The PCR reaction system is the same as in Example 1, and the annealing temperature is 44.3°C to 55.3° C., and the number of cycles is adjusted between 25 and 45. Refer to Example 1 for specific operations and determination methods.
[0067] 2. Experimental results
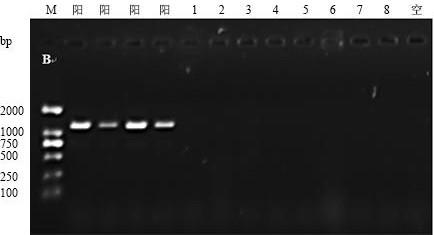
[0068] The results showed that the optimal annealing temperature for PCR using TTF-2 / TTR-2 (its nucleotide series as shown in SEQ ID NO: 1-2) as a primer was 52.3°C ( figure 2 ); the number of cycles is 25. Among them, "1~12" represent the annealing temperature of 44.3°C, 45.3°C, 46.3°C, 47.3°C, 48.3°C, 49.3°C, 50.3°C, 51.3°C, 52.3°C, 53.3°C, 54.3°C, 55.3°C, "13" represents a blank control.
Embodiment 3
[0069] Embodiment 3 a kind of clostridial disease detection kit
[0070] 1. Kit composition
[0071] The composition of the kit in this embodiment includes: primer TTF-2 / TTR-2 (the nucleotide series of which is shown in SEQ ID NO: 1-2), sample DNA extract, PCR reaction reagent and positive control.
[0072] Among them, the sample DNA extraction solution is an independently packaged fecal DNA extraction solution, and the specific composition is: 140 mL of ASL Buffer, 1.4 mL of proteinase K, 33 mL of Buffer AL, 19 mL of Buffer AW1, and 13 mL of Buffer AW2, 12 mL of Buffer AE, 10 mL of 96%-100% alcohol.
[0073] The PCR reaction reagents are: 500 μL Premix Ex Taq Version 2.0 (Loading dyeMix) for 40 reactions.
[0074] Primer TTF-2 / TTR-2: its nucleotide sequence is shown in SEQ ID NO: 1-2, and its concentration is 50 pmol / μL.
[0075] The positive control is Clostridium welchii genomic DNA, and its function is to compare PCR products and monitor whether the PCR operation proces...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com