Specific marker Nagorno-Karabakh deformation microsporidia nucleic acid probe and fluorescence in-situ hybridization detection method thereof
A fluorescence in situ hybridization and nucleic acid probe technology, which is applied in the field of in situ hybridization detection of N. naga proteus in silkworm parasitic cyst tissue, can solve the problem that the FISH labeling method of N. naga proteus has not been invented yet, and the structure cannot be cultured in vitro , Weak research foundation and other issues, to achieve the effect of intuitive and reliable results, easy operation, simple and convenient operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
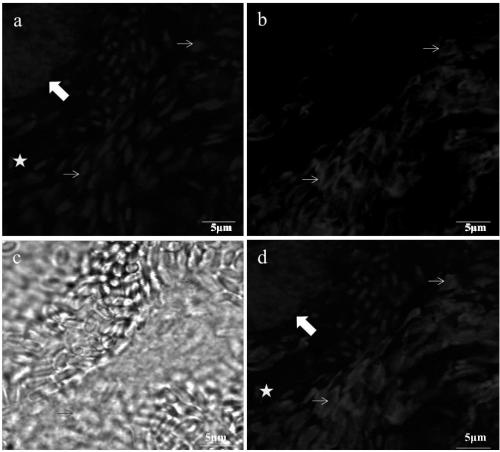
[0032] Embodiment 1, probe design
[0033] According to the DNA sequence of VnBM ribosomal RNA large subunit (LSU rRNA) and its secondary structure model, combining with FISH probes requires the design and screening of FISH probes. The requirements for the FISH probe are: the length is about 18nt, and the Tm value is 48-60°C. If the Tm value is greater than 60°C or less than 48°C, the length of the probe can be adjusted appropriately. According to the above principles, a nucleic acid probe was designed, the sequence of which is as follows:
[0034] VnLSU-V1: 5'-gtattctattacgaccttc-3' (SEQ ID NO.1);
[0035] The nucleic acid probe designed by the technical solution can specifically bind the ribosomal RNA outside the nucleus, while the general probe can only bind the DNA in the nucleus and cannot achieve the purpose of distinguishing the microsporidia of silkworm in different stages.
[0036] The Naga proteus nucleic acid probe is coupled with different fluorescent markers as ...
Embodiment 2
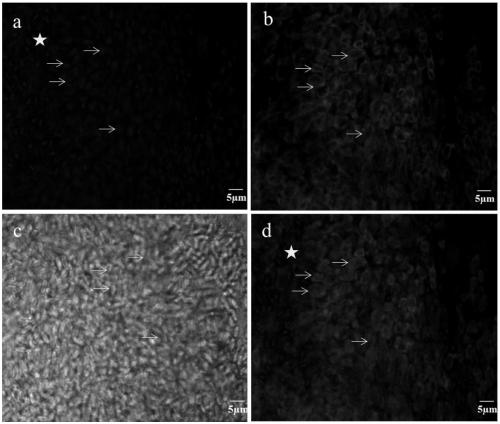
[0039] Embodiment 2, the method and result of VnLSU-V1 in situ hybridization experiment
[0040]1) Preparation of silkworm parasitic bursa tissue infected with VnBM
[0041] Bombyx mori were reared to the third instar, and the Naka proteus was evenly spread on the mulberry leaves, and 10 5 The amount of spores / silkworms was added to feed silkworms, and the growth and pathological changes of silkworms were observed and recorded. After the silkworm enters the fifth instar, the silkworm is dissected and the material obtained is the parasitic sac formed in the muscle cells of the rear midgut of the silkworm. Use tweezers to tear off the white tumor-like parasitic sac particles in the rear part of the silkworm midgut and place them in 1.5mL centrifuge tubes, take 6 tubes, fix 3 tubes with 4% paraformaldehyde, fix 3 tubes with 2.5% glutaraldehyde, and paste Label and store at 4°C.
[0042] 2) FISH probe labeling of hybridized VnBM in silkworm parasitic bursa
[0043] Put the par...
Embodiment 3
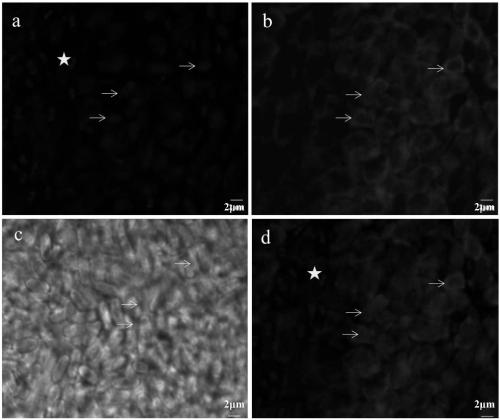
[0051] Embodiment 3, the method and result of VnLSU-V1-Cy3 in situ hybridization experiment
[0052] 1) Preparation of silkworm parasitic bursa tissue infected with VnBM
[0053] Bombyx mori were reared to the third instar, and the Naka proteus was evenly spread on the mulberry leaves, and 10 5 The amount of spores / silkworms was added to feed silkworms, and the growth and pathological changes of silkworms were observed and recorded. After the silkworm enters the fifth instar, the silkworm is dissected and the material obtained is the parasitic sac formed in the muscle cells of the rear midgut of the silkworm. Use tweezers to tear off the white tumor-like parasitic sac particles in the rear part of the silkworm midgut and place them in 1.5mL centrifuge tubes, take 6 tubes, fix 3 tubes with 4% paraformaldehyde, fix 3 tubes with 2.5% glutaraldehyde, and paste Label and store at 4°C.
[0054] 2) FISH probe labeling of hybridized VnBM in silkworm parasitic bursa
[0055] Put th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com