Quantitative detection kit for hepatitis B virus nucleic acid
A hepatitis B virus and detection kit technology, applied in the fields of molecular biotechnology and gene detection, to achieve the effects of high-sensitivity real-time fluorescent quantitative detection, optimization of experimental system, and improvement of amplification efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Embodiment 1 Novel highly sensitive hepatitis B virus nucleic acid quantitative detection kit
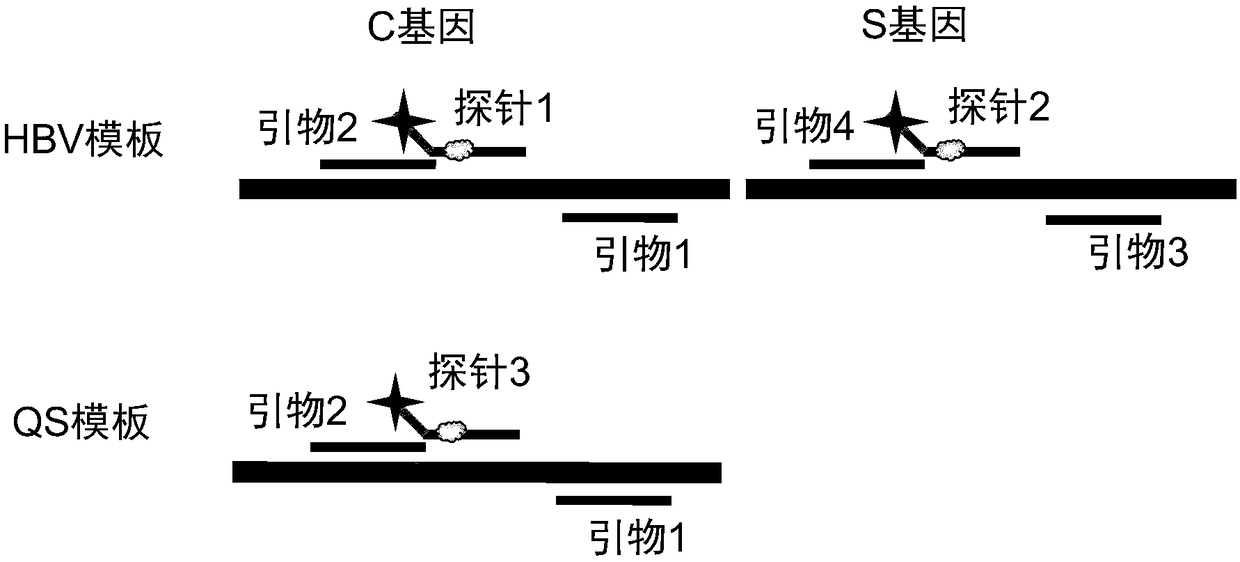
[0031] Establish a PCR combined with nucleic acid invasion reaction method for real-time quantitative detection of HBV DNA, the principle is as follows figure 1shown. The target template is amplified by designing a pair of PCR primers. In order to increase the binding efficiency of the primers and the template to be tested, in particular, LNA base modifications are added to the primers to further improve the amplification efficiency of the HBV template. For the amplification product, use primer 2 as the upstream probe to design a fluorescent downstream probe, which can anneal and hybridize with the template to form a single-base overlapping intrusion structure, which can be specifically recognized by Afu enzyme, thereby cutting the downstream probe and The template is complementary to the chemical bond of the first base, which produces a fluorescent signal. Since the Tm val...
Embodiment 2
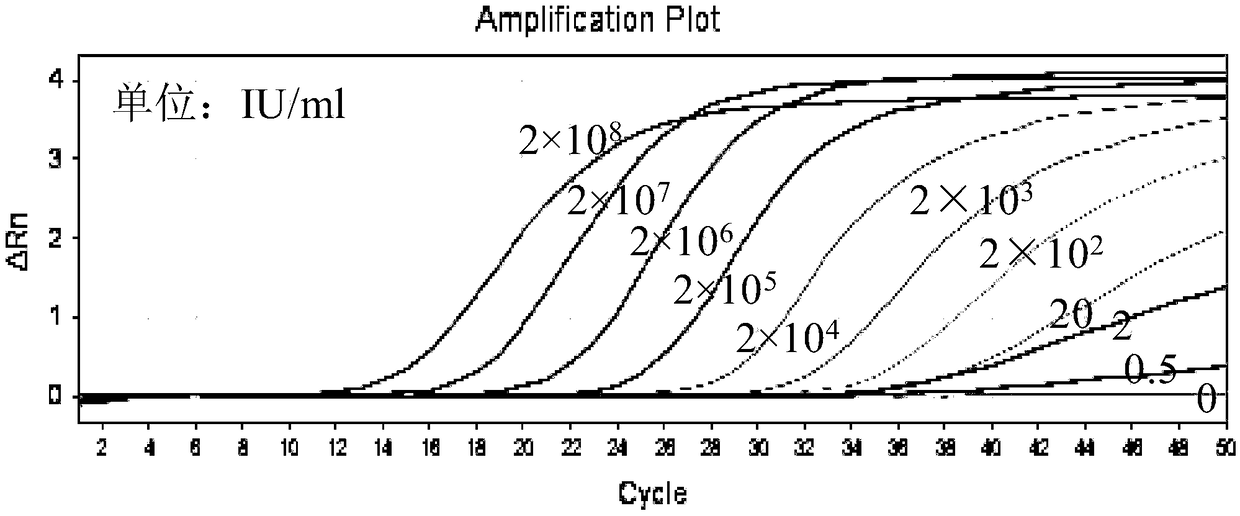
[0044] The sensitivity of embodiment 2 hepatitis B virus nucleic acid quantitative detection kits
[0045] In this example, the hepatitis B virus nucleic acid quantitative detection kit described in Example 1 was used to detect different concentrations of HBV DNA templates to verify the sensitivity of the detection method of the present invention.
[0046] Reaction conditions:
[0047] The 50 μL amplification system includes: 5 μL 10×buffer, 250 μmol / L dNTP, 500 nmol / L PCR upstream and downstream primers (SEQ ID NO.1 to SEQ ID NO.4), 250 nmol / L probe (SEQ ID NO.5 to SEQ ID NO. ID NO.7), 3U TaqDNA polymerase, 40U Afu enzyme and 20 μL plasma to extract HBV template DNA. HBV template concentrations were: 2×10 8 , 2×10 7 , 2×10 6 , 2×10 5 , 2×10 4 , 2×10 3 , 2×10 2 , 20, 2, 0.5 and 0IU / ml (QS quantitative template is 10 3 copy / μL), add water to make up to 50 μL. Amplification reaction program: 95°C, 3min; 95°C for 20s, 67°C, 30s, 70°C, 20s, 5 cycles of pre-amplification;...
Embodiment 3
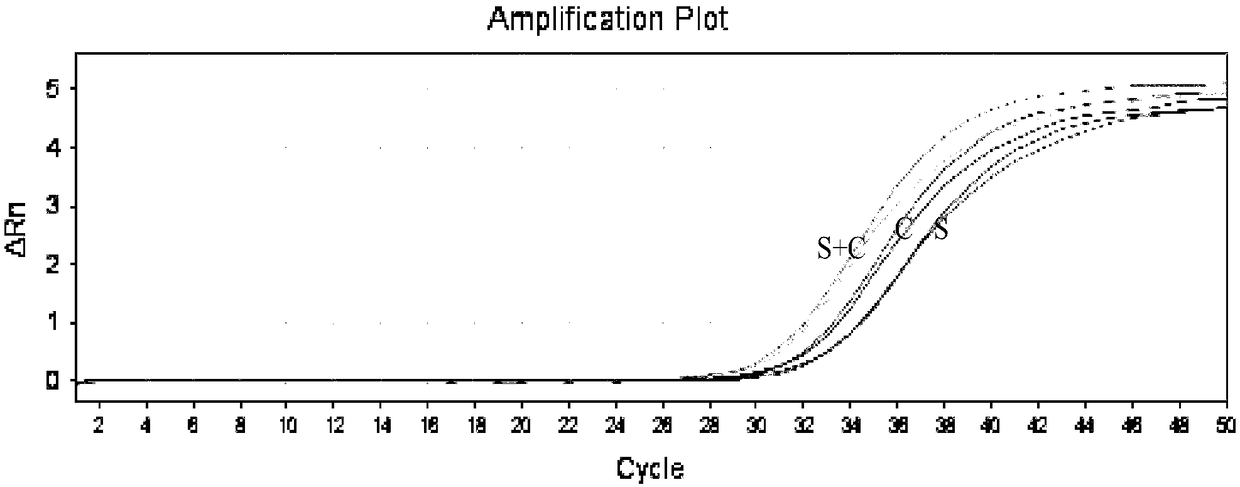
[0049] The quantitative limit of embodiment 3 hepatitis B virus nucleic acid quantitative detection kits
[0050] This embodiment uses the hepatitis B virus nucleic acid quantitative detection kit described in Example 1 to detect HBV standard templates of different concentrations mixed in human serum samples, which is used to verify the detection method of the present invention when detecting simulated samples Quantitation limit detection situation.
[0051] Reaction conditions:
[0052] The 50 μL amplification system includes: 5 μL 10×buffer, 250 μmol / L dNTP, 500 nmol / L PCR upstream and downstream primers (SEQ ID NO.1 to SEQ ID NO.4), 250 nmol / L probe (SEQ ID NO.5 to SEQ ID NO. ID NO.7), 3U TaqDNA polymerase, 40U Afu enzyme and 20 μL plasma to extract HBV standard template DNA. HBV standard template concentrations are: 2×10 7 , 2×10 6 , 2×10 5 , 2×10 4 , 2×10 3 , 2×10 2 , 20 and 2IU / ml (QS quantitative template is 10 3 copy / μL), add water to make up to 50 μL. Amplif...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com