RPA primer for detecting mycoplasma ovipneumoniae
A technology of mycoplasma pneumoniae and sheep, applied in the direction of microorganism-based methods, DNA/RNA fragments, microorganisms, etc., can solve the problems of high nutritional requirements of the medium, difficult growth, poor specificity, etc., and achieve accurate detection results, good repeatability, The effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Embodiment 1: be used to detect the establishment of the RPA method of ovine mycoplasma pneumoniae
[0024] 1. Design and synthesis of primers
[0025] The specificity of the primers is an important factor in the method established in this experiment. According to the p80 gene sequence of KM435069.1 registered in GenBank, primers were designed by using Oligo 7 software, and their specificity was preliminarily verified by comparison and analysis by BLAST software. Primer synthesis was completed by Sangon Bioengineering (Shanghai) Co., Ltd.
[0026] The nucleotide sequence of the RPA primer is:
[0027] Primer upstream P1: 5'- CTTAATGATGAGCGTAGTAGATACCAAACCAC -3',
[0028] Downstream primer P2: 5'-GCTGAGGTCGGATTTGGACTAACAATATTTG-3'.
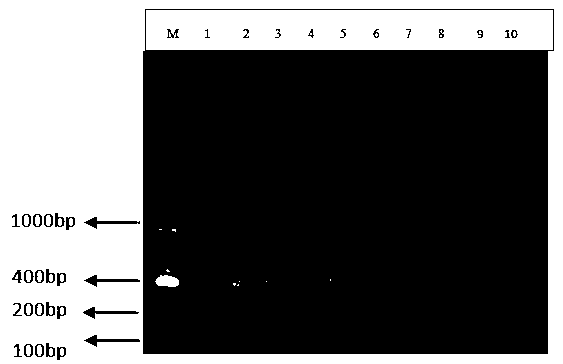
[0029] The size of the amplified fragment was 353 bp.
[0030] 2. Preparation of positive standard
[0031] Using the extracted DNA as a template, take 1 mL of Mycoplasma ovis pneumoniae culture solution and add it to a 1.5 mL centrifu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com