Single-stranded DNA aptamer specifically recognizing tobramycin and application thereof
A tobramycin and aptamer technology, applied in the field of single-stranded DNA aptamers, can solve the problems of affinity, sensitivity reduction, affinity reduction, destruction of secondary structure, etc., and achieve high sensitivity, high affinity and strong specificity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: Tobramycin ssDNA aptamer truncation
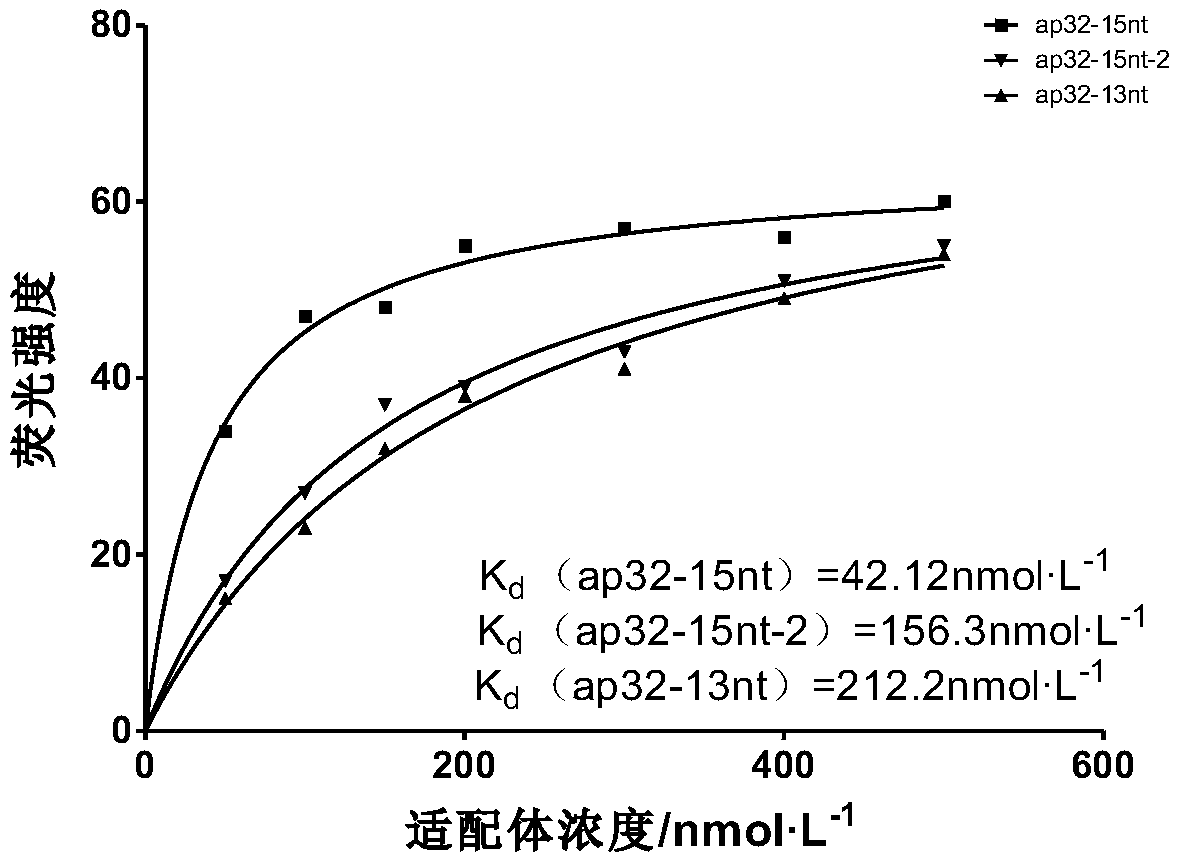
[0025] The aptamer ap32-34nt (Kd=58.92nmol·L) obtained in previous research in the laboratory -1 , its nucleotide sequence is shown in SEQ ID NO.4, which is 5'-CGTCGACGGATCCATGGCACGTTATAGGTCGACG-3') on the basis of the secondary structure (for the aptamer ap32-34nt, refer to the large paper "Tobramycin-specific single-stranded DNA Screening of aptamers and their sequence optimization and application research" Zhang Yuhong, Jiangnan University), truncated and optimized, and obtained several sets of aptamer sequences, as shown in Table 1:
[0026] Table 1 Aptamers and corresponding nucleotide sequences
[0027] Aptamer
Nucleotide sequence (SEQ ID NO.1~3)
ap32-15nt
5'-GACTAGGCACTAGTC-3'
ap32-13nt
5'-GACGGGCACCGTC-3'
ap32-15nt-2
5'-GACGTGGCACACGTC-3'
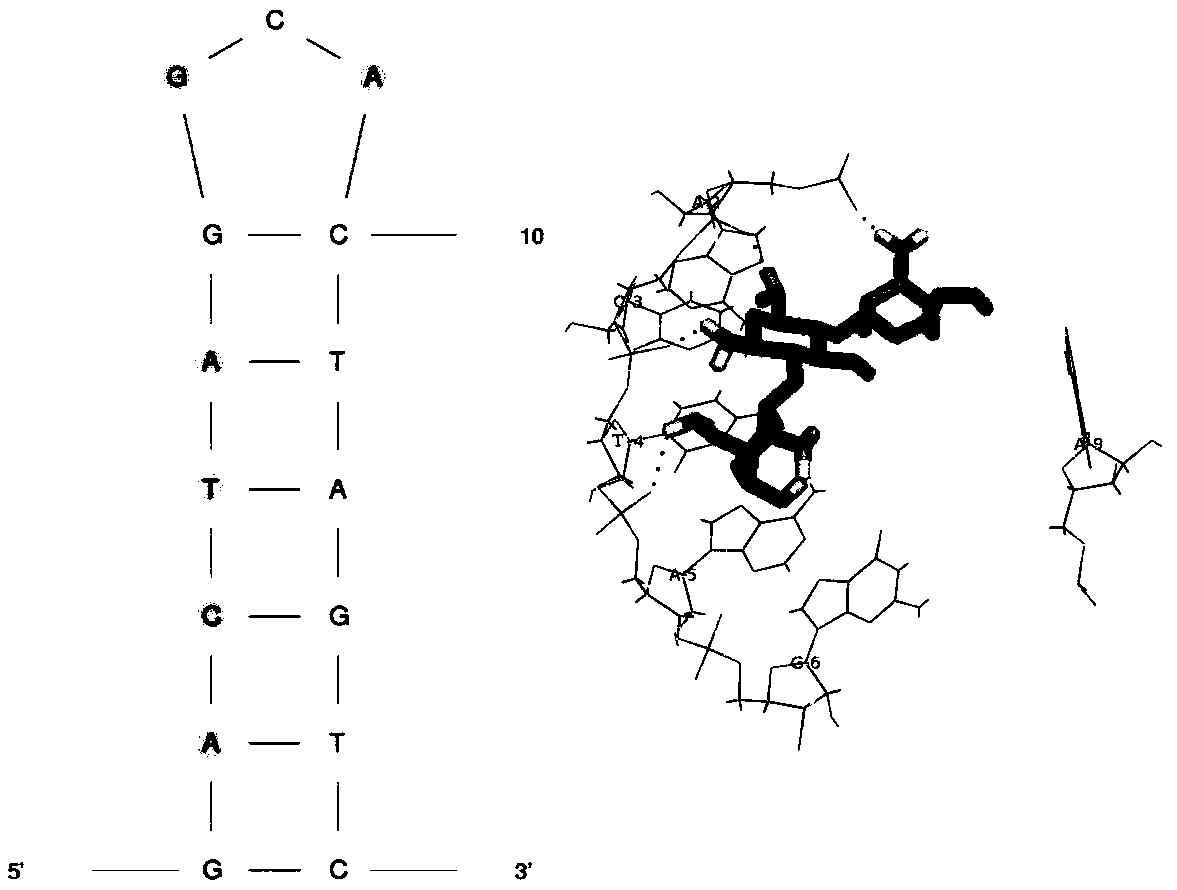
[0028] To analyze the stem-loop structure, the dissociation constant (K d ) determination, the specific steps are as follo...
Embodiment 2
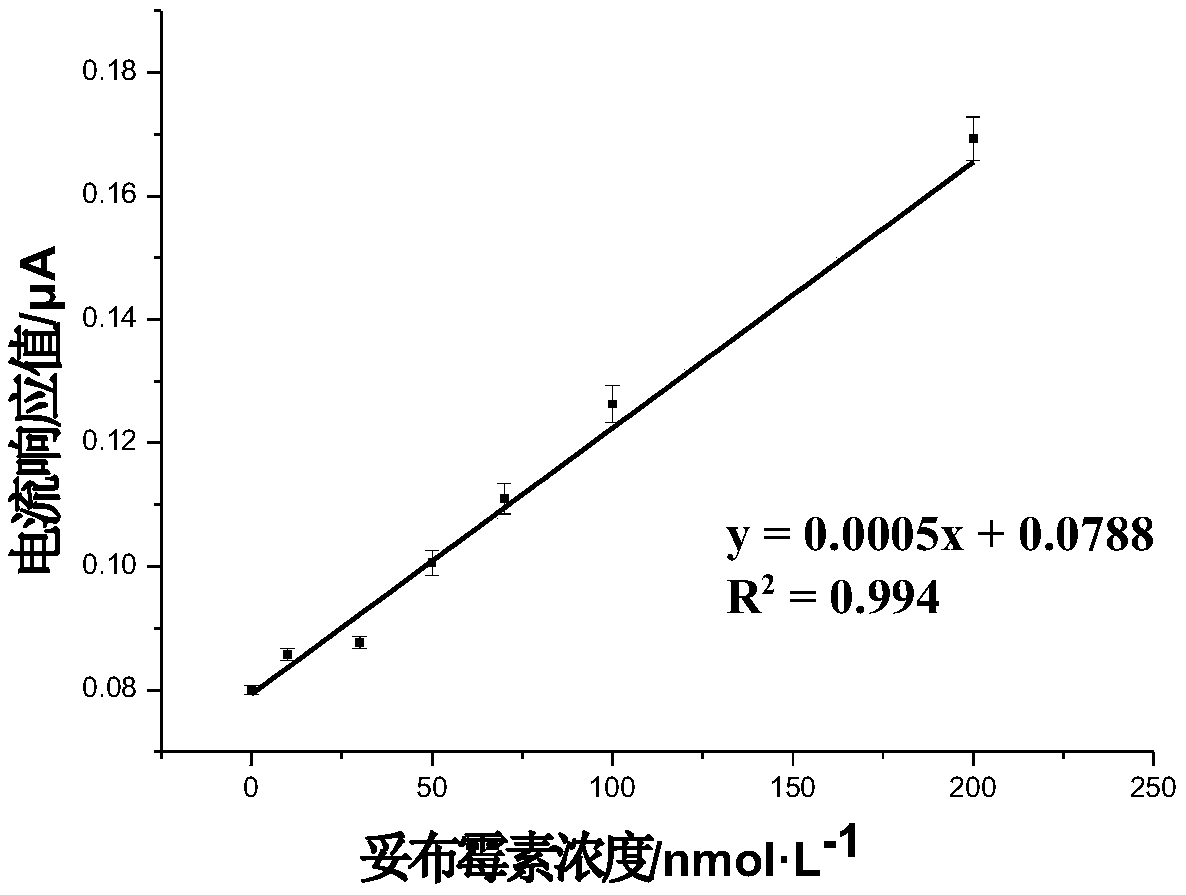
[0042] Example 2: Aptamer ap32-15nt electrochemical method to detect tobramycin
[0043] The designed hairpin structure sequence is (SEQ ID NO.5): 5'-AAAAAAGACTAGGCACTAGTCAAAAAAACCCCCGATCCTAGTCTTTCCC-3'; the italic part is the truncated aptamer sequence.
[0044] The designed signal transduction probe sequence is (SEQ ID NO.6): 5'-GCGAAAAAAGCG-(CH 2 ) 6 -HS-3', the 3' end of the probe is modified with a sulfhydryl group to self-assemble onto the gold electrode surface.
[0045] The designed primer sequence is (SQE ID NO.7): 5'-AAAGACTAGGA-3'
[0046] (1) Sequence design of hairpin structure (Hp), signal transduction probe, and primer; and preparation concentrations were 10 μmol L -1 , 1 μmol L -1 , 10 μmol L -1 .
[0047] (2) Trichlorohexaammine ruthenium ([Ru(NH 3 ) 6 ] 3+ ) solution: use 10mmol·L -1 Tris-HCl prepared at a concentration of 10 μmol L -1 .
[0048] (3) The hairpin structure (Hp) in step (1) was heated at 95° C. for 5 minutes and placed in a hot wate...
Embodiment 3
[0056] Embodiment 3: the detection of tobramycin by gold gel colorimetric kit
[0057] (1) Solution A: AuNPs were prepared by trisodium citrate reduction method and concentrated 5 times;
[0058] (2) Solution B: tobramycin aptamer ap32-15nt, prepared at a concentration of 150nmol L -1 ;
[0059] (3) Liquid C: NaCl solution, the preparation concentration is 120mmol L -1 ;
[0060] (4) Take 50 μL of solution A in step (1) and 100 μL of solution B in step (2) and react for 1 hour at room temperature in the dark;
[0061] (5) Add different concentrations of tobramycin to step (4) successively, and react in the dark for 50 minutes;
[0062] (6) Add 50 μL of solution C in step (3) to the solution obtained in step (5), observe the color change of each centrifuge tube, perform spectral characterization with a spectrophotometer, and draw a graph of the relationship between the absorbance of the AuNPs solution and the concentration of tobramycin.
[0063] In the presence of differe...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com