Method for rapidly determining genome size of sturgeons by flow cytometry
A flow cytometry and rapid determination technology, which is applied in the field of determination of the genome size of sturgeon species, can solve the problems of complicated processing technology, infection risk of incisions, increased feeding costs, etc., and achieve the effect of rapid determination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
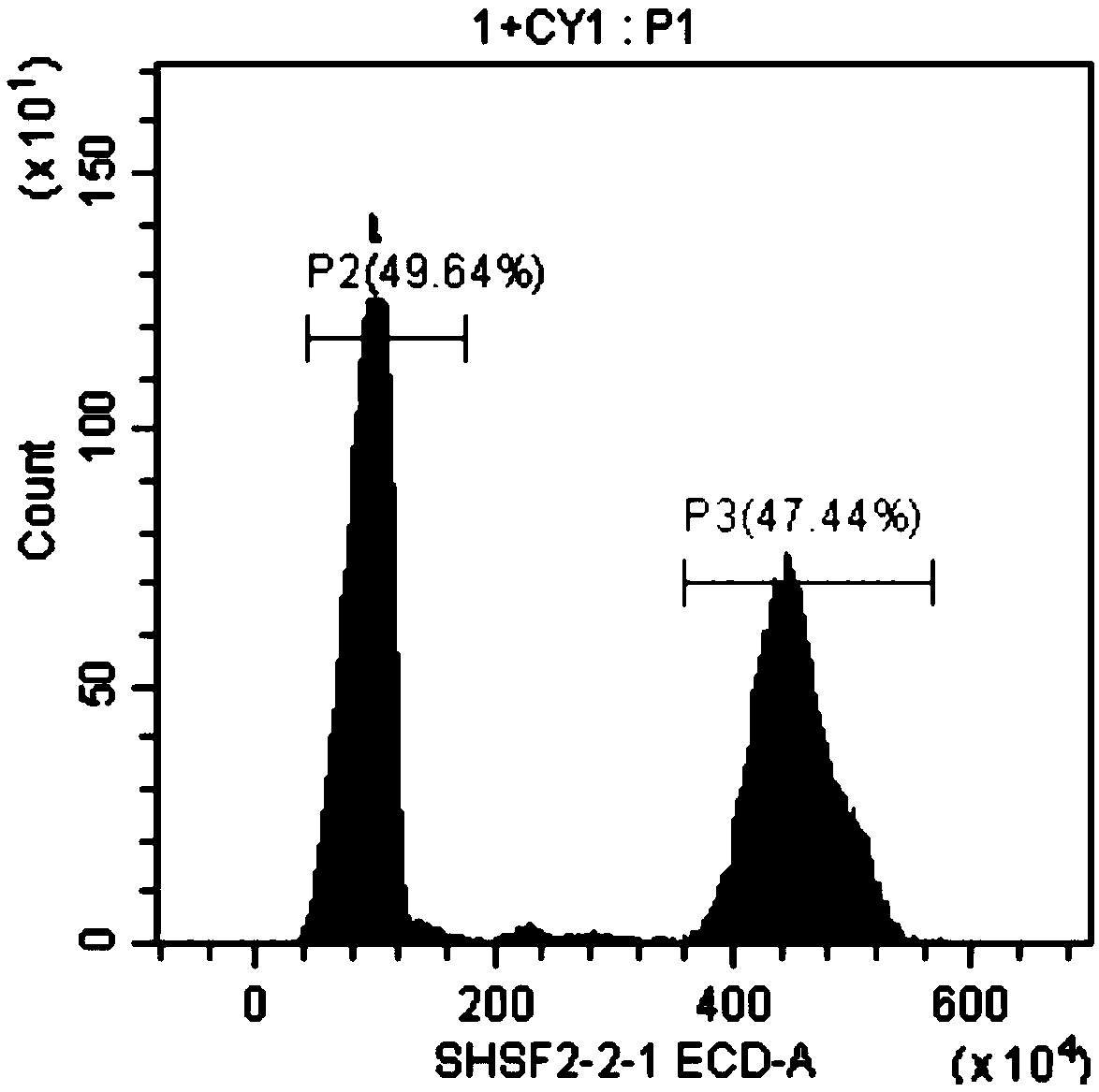
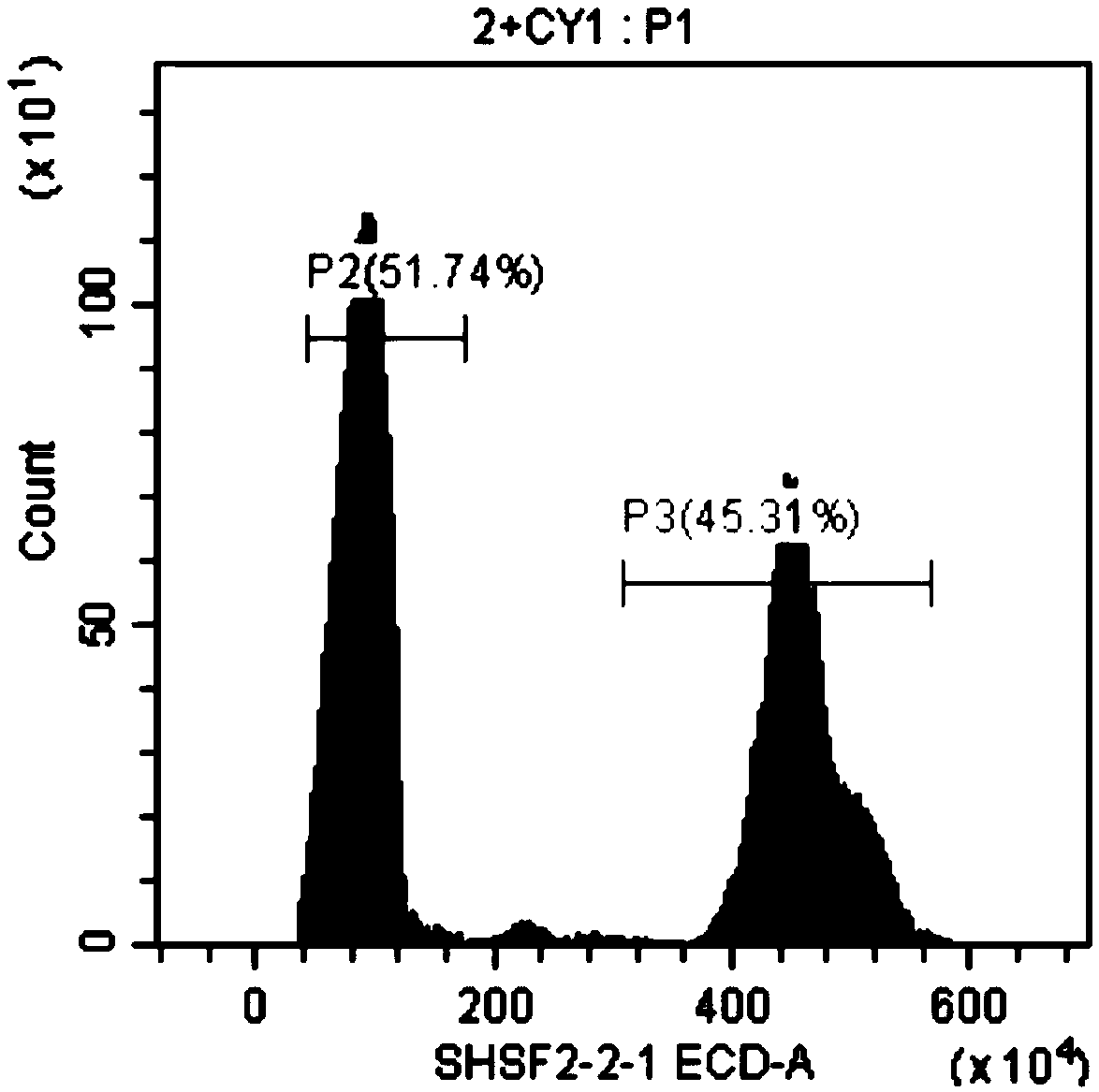
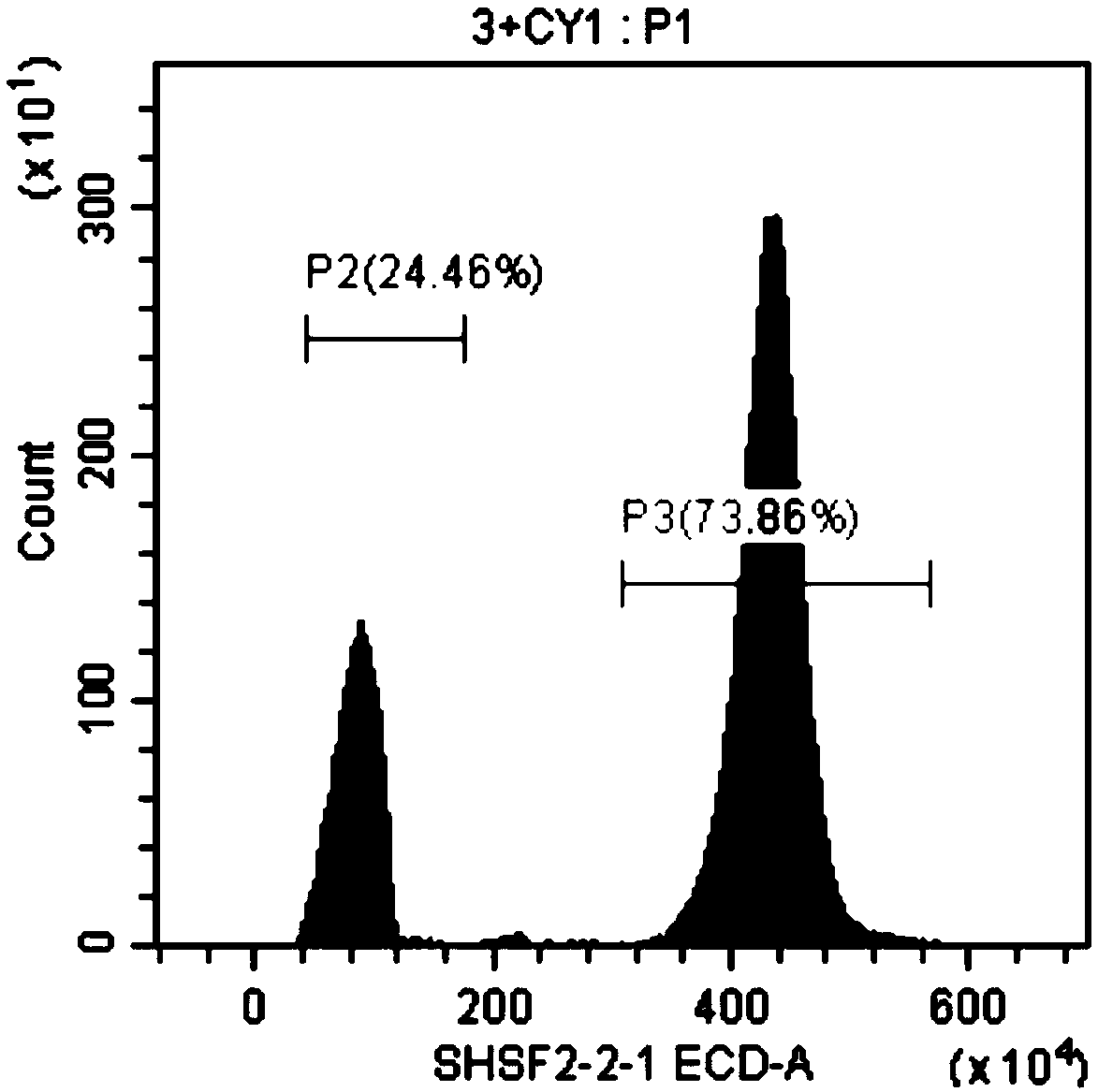
[0047] Embodiment 1, the method for rapidly measuring the genome size of sturgeon by flow cytometry, carries out the following steps successively:
[0048] 1. Grass carp, a carp with known genome size, was selected as a standard control. The genome size of grass carp is about 1.07 Gb. Steps 1) to 7) of the following operations are performed on the grass carp and the sturgeons to be tested (female Acipenser dauribus, male Acipenser dauribus, female Acipenser schneidere, male Acipenser schneidere, female Russian sturgeon and male Russian sturgeon):
[0049] 1) Take blood from the tail vein, take 0.5ml of blood, and store it in EDTA anticoagulant tubes.
[0050] 2) Take fresh blood or anticoagulated blood and centrifuge at 1500rpm for 5min. Wash twice with PBS buffer (remove the washing solution after each wash, the same below), add 0.5ml PBS buffer to resuspend red blood cells; obtain red blood cell suspension I.
[0051] 3) Take 50 μl of the erythrocyte suspension I obtained ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com