Cancer genome and non-specific gene tag-based large-scale drug relocation method
A gene and labeling technology, applied in the field of large-scale drug repositioning, can solve problems such as inability to be used in clinical treatment, interference of expression profile tags, and inability to implement in humans
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach
[0065] As a specific embodiment of the present invention, the data is obtained from the Connectivity Map (CMap) database, preferably, the drug treatment expression profile tags are analyzed from the microarray chip of the CMap database, so as to be integrated with the core tags for analysis.
[0066] In a specific embodiment of the present invention, based on the drug repositioning method of the present invention, the core tags determined by the method of the present invention are compared with 3,546 sets of drug processing data, and 5,362,359 drug repositioning candidate results are determined, including 2,511,089 drug-gene interaction relationships predicted for unknown target drugs were obtained.
[0067] In the method of the present invention, after obtaining the drug repositioning candidate results, it also includes: performing further cell experiment / animal experiment verification on the function (potential therapeutic effect) determined after drug repositioning, so as to...
Embodiment 1
[0073] Example 1. Construction of core tags of key signaling pathway genes from cancer transcriptome
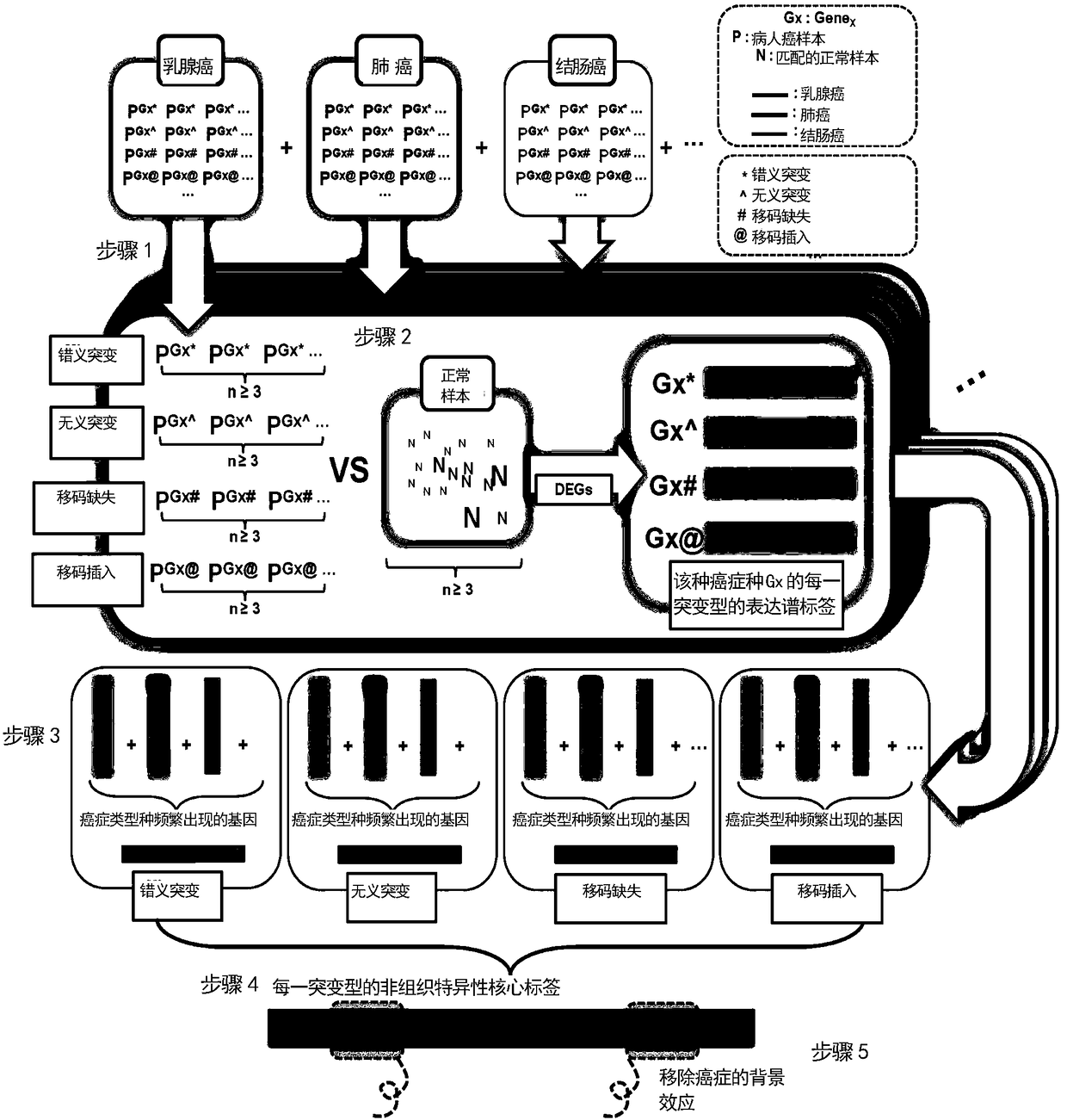
[0074] In order to construct the core tags of key signaling pathways, in this example, 4,895 key signaling pathway genes responsible for encoding receptors, signaling cascade factors and transcription factors were collected.
[0075] The TCGA database provides thousands of sets of high-quality transcriptome data for more than 20 cancer types, as well as tissue-type-matched normal control data. In this embodiment, the TCGA database is used as the basis for determining the core tags.
[0076] Based on the TCGA database, the inventor has designed 5 steps to realize the construction of core tags ( figure 1 ). For each cancer type, the patient sample group carrying a mutation type of a gene (missense mutation, nonsense mutation, frameshift mutation deletion or insertion, etc.) Control group comparison to find the expression profile signature of the gene in different mutation ty...
Embodiment 2
[0083] Example 2. Drug relocation based on gene core tags and drug processing transcriptome data
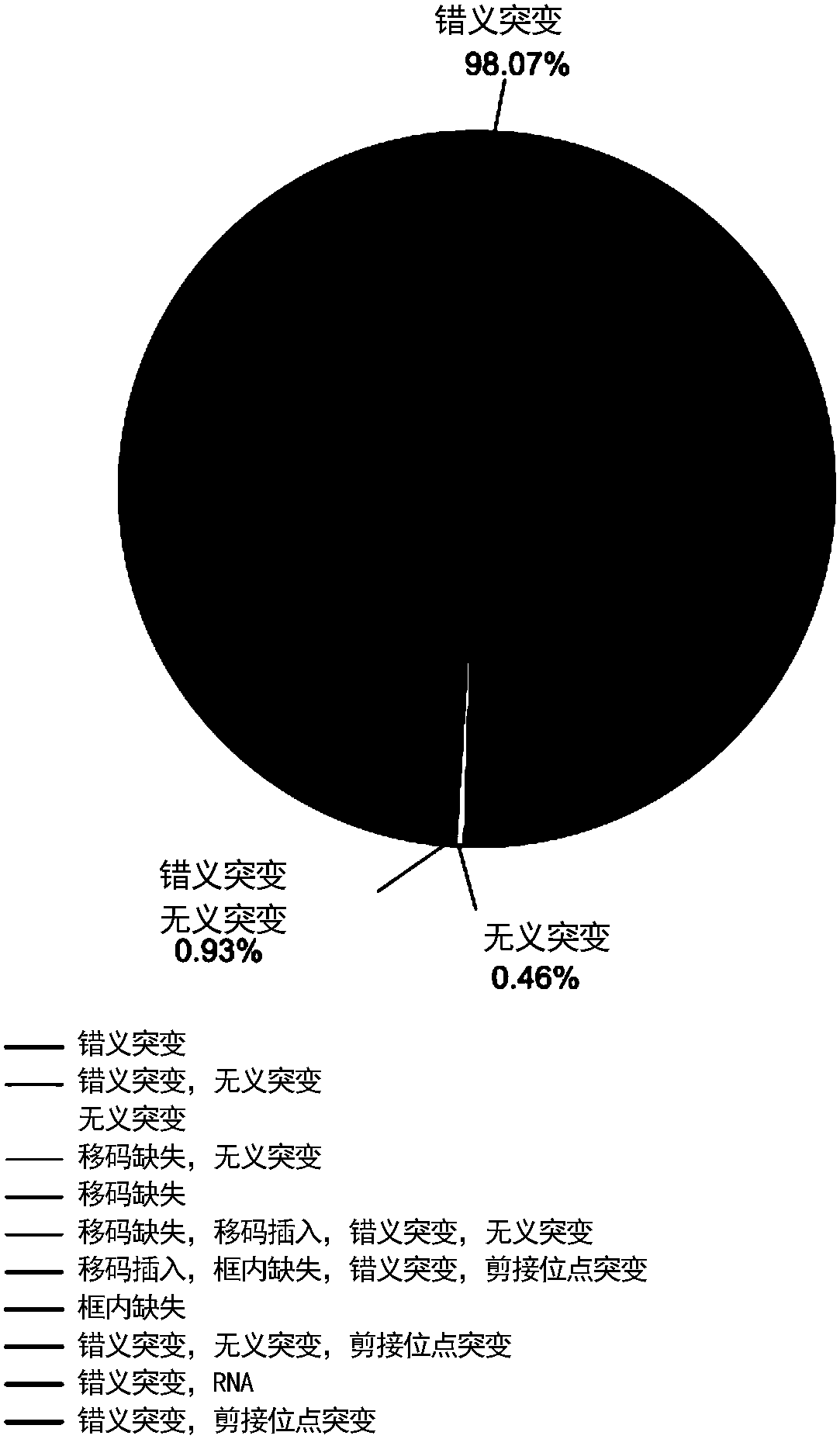
[0084] In order to apply the core tags to drug repositioning, the inventors analyzed 3,546 drug-processing expression profile tags from the microarray chip of the CMap database for integrated analysis with the core tags. The CMap database contains microarray data of 1,309 drugs in 3 human cancer cell lines. The different cell line data for each drug were analyzed separately. Because the inventors found that, although for the same drug, only 4% had more than 100 shared differentially expressed genes in different cell lines, 37% of the drugs did not have any shared differentially expressed genes in different cell lines. This indicates that the vast majority of drugs have different expression profile signatures in different cell lines.
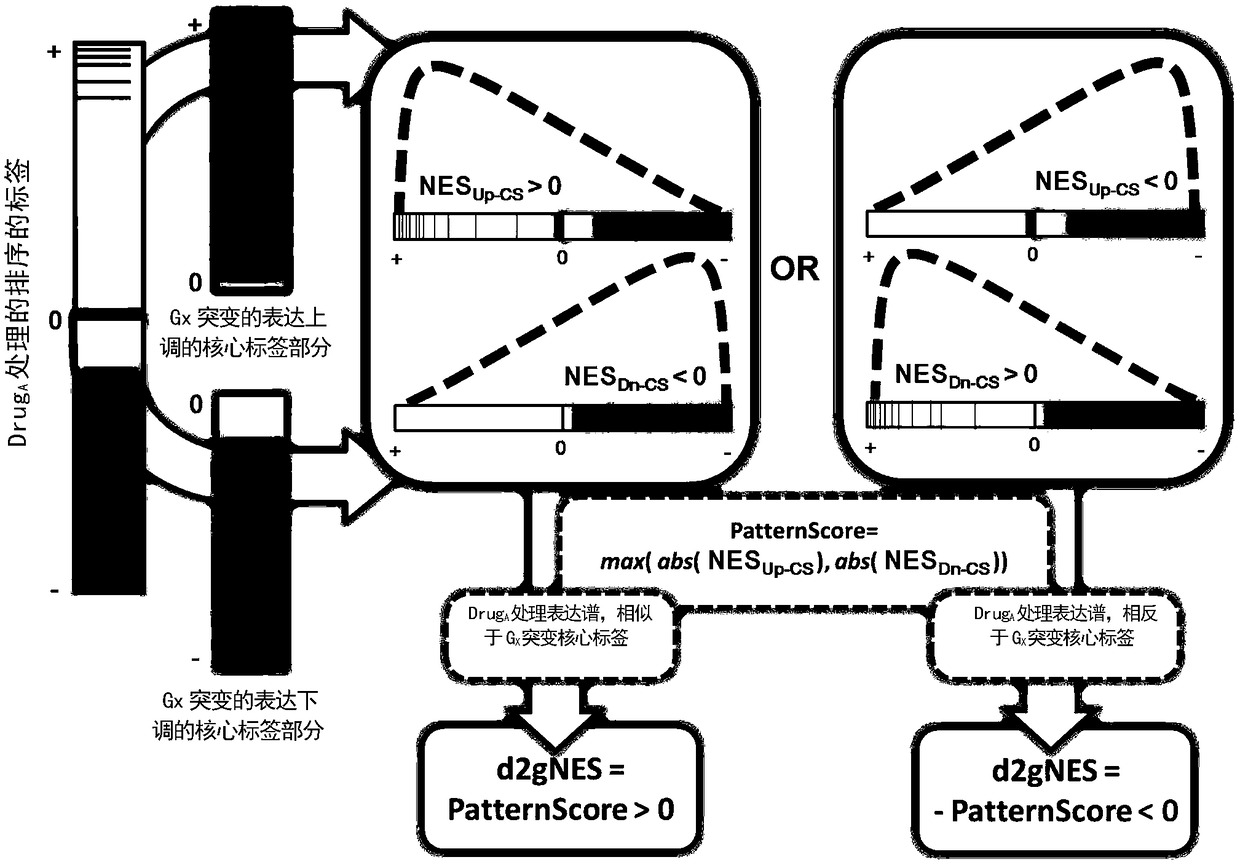
[0085] In order to integrate and analyze the core tags of gene mutations and drug treatment expression profile tags, the inventors evaluated the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com