sgRNAs that efficiently edit the porcine miR-17-92 gene cluster
A gene clustering and editing technology, applied in DNA/RNA fragments, genetic engineering, DNA preparation, etc., can solve problems such as limiting the breeding and application prospects of transgenic pigs, and achieve the effect of removing potential biological safety hazards and increasing safety.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] 1-1. Design of sgRNA sequence and construction of PX330 expression vector
[0026] Three sgRNA sequences targeting porcine miR-17-92 were designed and synthesized. The sgRNA sequences designed above were synthesized; the DNA sequences of the six single-stranded sgRNAs were annealed to form three oligonucleotide chains of sgRNAs targeting different positions of the porcine miR-17-92 3'-UTR; This oligonucleotide was then ligated into the pX330 plasmid vector.
[0027] The sequences of the three sgRNAs and the sequences of their action sites are:
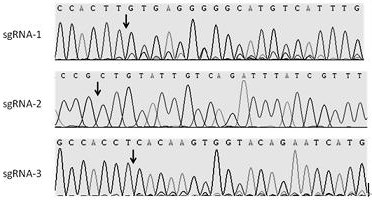
[0028] SgRNA-1 sequence: 5-atgattctgtaccacttgtg-3
[0029] The sequence of the sgRNA-1 action site: 5-CACAAGTGGTACAGAATCAT-3
[0030] SgRNA-2 sequence: 5-GCTGTATTGTCAGATTTATC-3
[0031] The sequence of the sgRNA-2 action site: 5-GATAAATCTGACAATACAGC-3
[0032] SgRNA-3 sequence: 5-attctgtaccacttgtgagg-3
[0033] The sequence of the sgRNA-3 action site: 5-CCTCACAAGTGGTACAGAAT-3
[0034] Among them, the present invention inv...
Embodiment 2
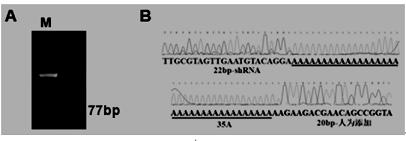
[0043] Construction of targeting vector for site-directed integration of shRNA
[0044] According to the screened high-efficiency sgRNA, the shRNA site-specific integration targeting vector (pLB-shRNA-KI-Donor) matching the sgRNA was designed and constructed. The main components of the targeting vector are: upstream homology arm, shRNA-EGFP gene, downstream homology Source arm and backbone vector for prokaryotic expression. The shRNA site-specific integration targeting plasmid and the screened sgRNA can specifically genetically modify the porcine miR-17-92 site, and then combine the method of fluorescence microscopy and PCR to easily analyze the recognition of foreign genes by the sgRNA Feasibility of site integration and expression ( figure 2 , schematic diagram of shRNA site-specific targeting plasmid vector).
Embodiment 3
[0046] Co-transfection of PX330 plasmid and pLB-shRNA-KI-Donor plasmid
[0047] Resuscitate PK-15-EGFP-KI cells, and when they are close to confluence, wash 2~3 times with DPBS, discard the supernatant, add electrotransfection buffer, and then press PX330 plasmid and pLB-shRNA-KI-Donor plasmid Add the ratio to the cells and the buffer solution, mix gently with a pipette, gently transfer the mixture into the electrode cup, and place the electroporation cup on the electroporation instrument for electric shock operation. After the electric shock was completed, the electro-rotator cup was left to stand for 10 minutes, and then the mixed solution in the electro-rotor cup was transferred to a cell culture dish. Finally, the cell culture dish was placed in a 37°C carbon dioxide incubator for cultivation. After 12 hours of incubation, the medium was changed.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com