Method for producing heavy chain aminocarboxylic acid
A transaminase gene and gene technology, applied in the field of production of medium-chain aminocarboxylic acids, can solve the problems of high yield production and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0029] In order to achieve the purpose of the present invention, the present invention provides a recombinant microorganism, wherein the fatty aldehyde dehydrogenase gene in the ω-oxidative metabolic pathway and the genes related to the β-oxidative metabolic pathway are deleted, and the ω-transaminase gene is also introduced therein.
[0030] In the present invention, the term "ω-oxidation" refers to a metabolic process in which the terminal methyl group of a fatty acid is oxidized to form a dicarboxylic acid, and the term "β-oxidation" refers to a metabolic process in which the carbon at the β position of the carboxyl group Atoms are oxidized to release acetyl-CoA, whereby fatty acids are gradually decomposed into fatty acids with a reduced number of carbon atoms by 2. The concepts of ω-oxidation and β-oxidation and the enzymes involved in this metabolic process are well known to those of ordinary skill in the field of biochemistry. For example, when fatty acids are used as s...
example 1
[0050] Example 1: Construction of a knockout cassette
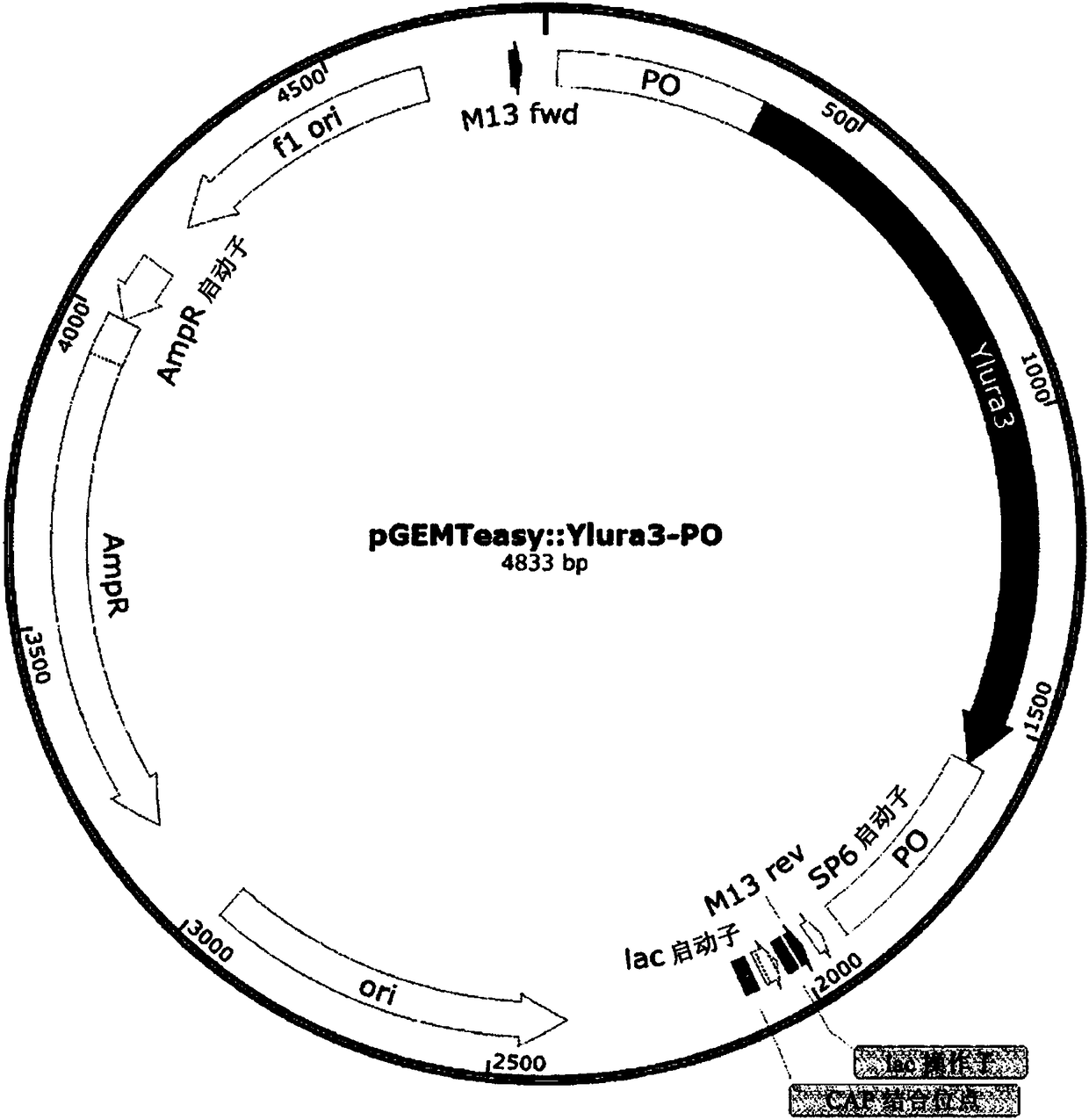
[0051] A vector was constructed containing the ura3 gene used as a selection marker for gene knockout for changing strains and a pop-up region for deletion of the ura3 gene after insertion of the knockout cassette ( image 3 ). A gene derived from Yarrowia was used as the ura3 gene, and the pop-up region for changing the strain had a total of four sequences, and two genes were referred to. Here, a glutamic acid production gene derived from Bacillus was used as one of the genes, and a His operon-related gene derived from Salmonella or the cloning vector pHUKH was used as the other gene. Table 1 below lists the primers and sequences used to construct the pop-out vector.
[0052] 【Table 1】
[0053]
[0054] like Figure 4 Knockout cassettes were constructed as indicated. First, PCR was performed on the homology region (HR) to be knocked out from the Yarrowia genomic DNA, and on the two 5' and 3' end fragments in th...
Embodiment 2
[0062] Embodiment 2: Construction of transduction vector
[0063] To insert an omega-transaminase into a Yarrowia strain, constructs such as Figure 5 vector shown. Table 4 lists the primers used for this purpose.
[0064] 【Table 4】
[0065]
[0066] with Figure 4 The transaminase cassette was constructed in the same manner except that when two PCR product fragments were obtained from the vector, the gene from the promoter to ura3 was amplified to construct the cassette. Primers used to construct the cassettes are listed in Table 5 below.
[0067] 【table 5】
[0068]
[0069] The gene sequences used to alter the recombinant microbial strains according to the present invention are listed in the sequence listing and summarized in Table 6.
[0070] 【Table 6】
[0071] Gene
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com