NY-ESO-1-targeting T cell receptor and uses thereof
A technology of NY-ESO-1 TCR and cell receptors, which is applied in the field of T cell receptors targeting NY-ESO-1, and can solve problems affecting the killing function of T cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Example 1: Determination of NY-ESO-1 TCR gene sequence
[0067] The sequence information of human NY-ESO-1 TCR α chain, P2A, NY-ESO-1 TCR β chain gene sequence information was searched from the NCBI website database, and these sequences were codon optimized on the website http: / / sg.idtdna.com / site, It is guaranteed that it is more suitable for human cell expression under the condition that the encoded amino acid sequence remains unchanged.
[0068] See (SEQUUNCE ID NO.1-2) for each amino acid and gene sequence information.
[0069] Ligate the above sequence according to the human NY-ESO-1 TCRα chain, P2A, and NY-ESO-1 TCRβ chain in sequence, and introduce enzyme cutting sites at the sequence junction to form a complete NY-ESO-1 TCR gene sequence information .
Embodiment 2
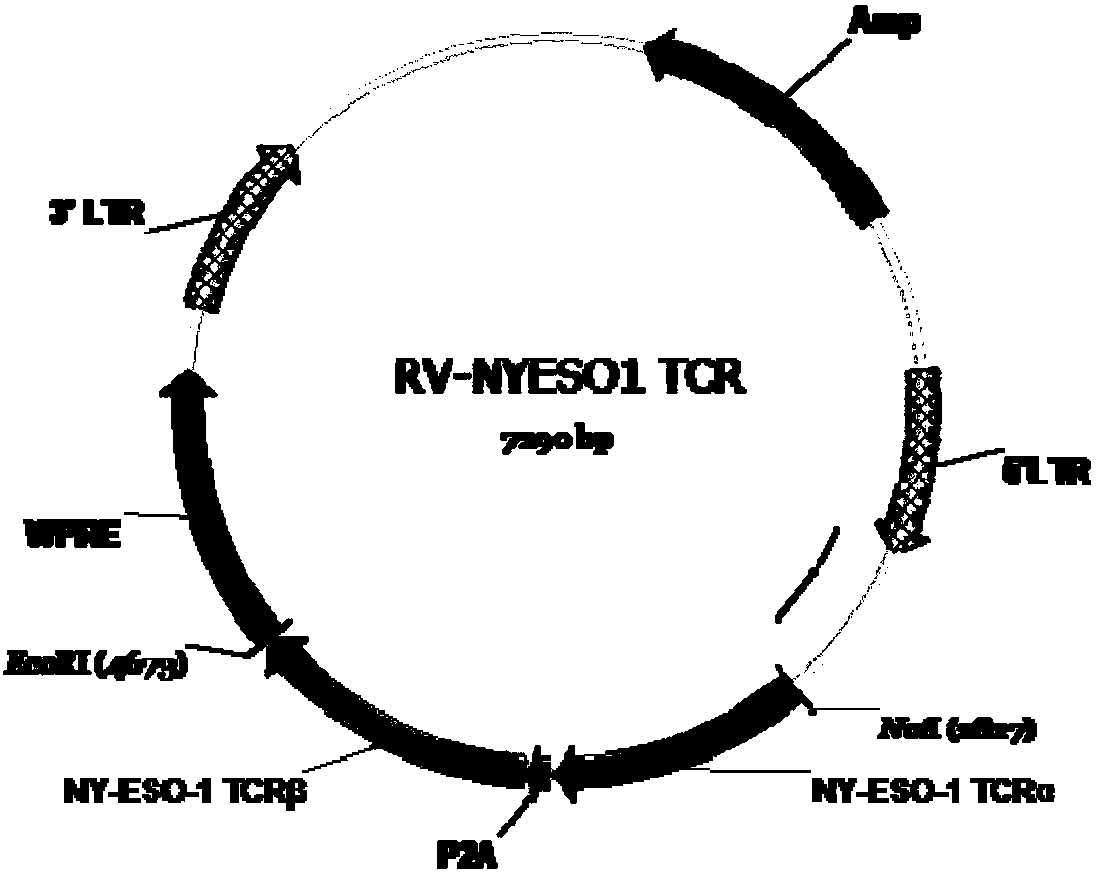
[0070] Embodiment 2: the construction of the viral vector comprising the nucleic acid sequence of TCR molecule
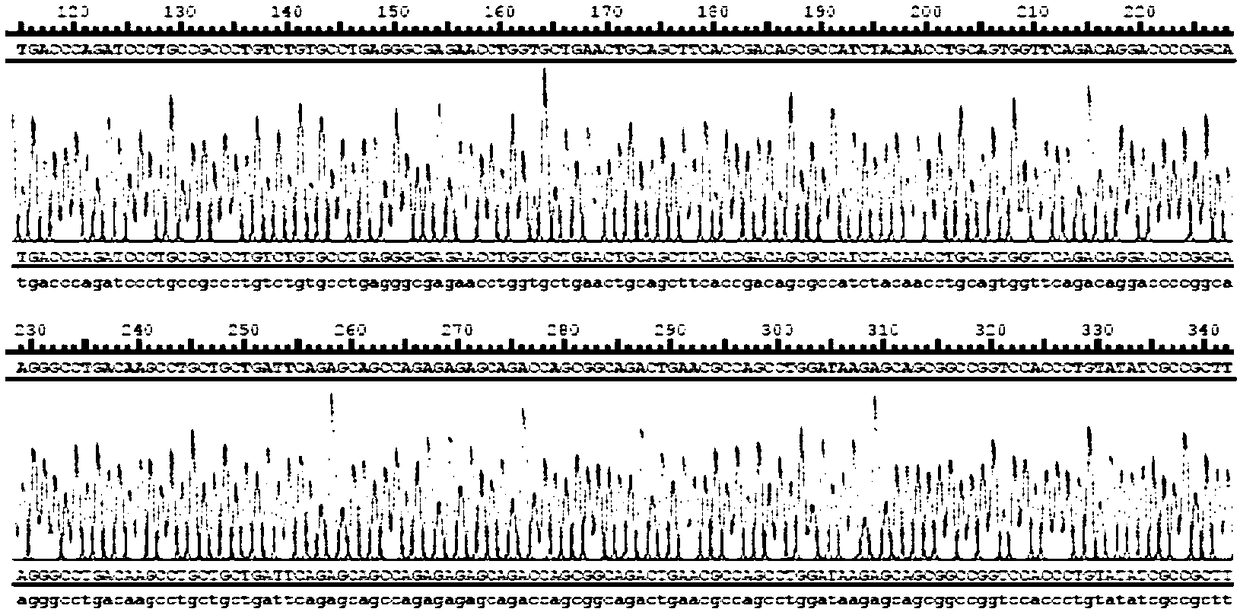
[0071] The nucleotide sequence of the TCR molecule prepared in Example 1 was double digested with NotI (NEB) and EcoRI (NEB), connected and inserted into the NotI-EcoRI site of the retroviral RV vector through T4 ligase (NEB), and transformed into When the competent E.coli (DH5α) was obtained, the recombinant plasmid was sent to Shanghai Sangon Biotechnology Co., Ltd. for sequencing, and the sequencing result was compared with the fitted NY-ESO-1TCR sequence to verify whether the sequence was correct. The sequencing primers are:
[0072] Sense sequence: AGCATCGTTCTGTGTTGTCTC
[0073] Antisense sequence: TGTTTGTCTTGTGGCAATACAC
[0074] After the sequencing was correct, the plasmid was extracted and purified using the plasmid purification kit from Qiagen, and the purified plasmid was transfected into 293T cells by the plasmid calcium phosphate method for retrovirus ...
Embodiment 3
[0076] Example 3: Retroviral Packaging
[0077] 1. On the first day, the 293T cells should be less than 20 passages and not overgrown. Take 0.6×10 6 Cells / ml plated, add 10ml DMEM medium to a 10cm dish, mix the cells well, and culture overnight at 37 degrees;
[0078] 2. On the second day, the 293T cell confluency reaches about 90% for transfection (usually about 14-18 hours after plating); prepare the plasmid complex, the amount of various plasmids is 12.5ug for Retro backbone (MSCV), and 12.5ug for Gag-pol 10ug, VSVg is 6.25ug, CaCl 2 250ul,H2 O is 1ml and the total volume is 1.25ml; add HBS equal to the volume of the plasmid complex in another tube, and vortex for 20 seconds while adding the plasmid complex. Gently add the mixture to the 293T dish along the side, incubate at 37 degrees for 4 hours, remove the medium, wash with PBS, and re-add the preheated fresh medium;
[0079] 3. Day 4: 48 hours after transfection, collect the supernatant and filter it with a 0.45um ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com