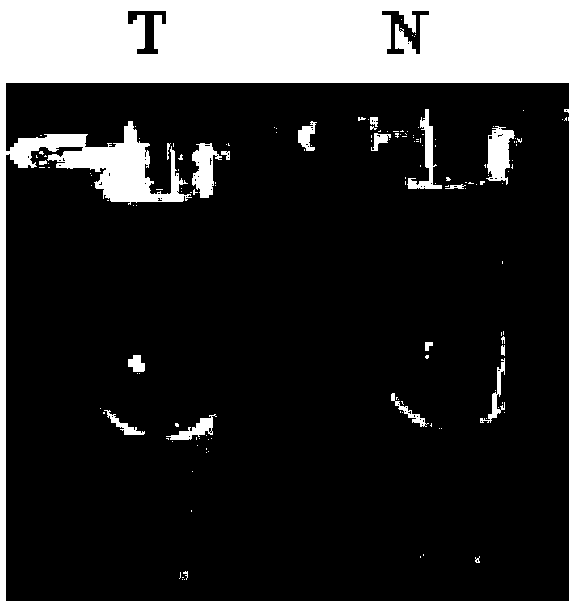Direct-spread RPA (recombinase polymerase amplification) on-site visibility detection method
A detection method and amplification reaction technology, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0099] Example 1 is used to detect the acquisition of the RPA primer set of transgenic maize TC1507
[0100] Primers are designed according to the conserved sequence of transgenic maize TC1507 found in GMDD (GMO Detection method Database). Due to the immature development of RPA technology, there is no special software for designing primers like PCR or LAMP. Therefore, the primer design of RPA requires manual Manually designed, further verified in experiments, and screened out the most suitable primer pairs.
[0101] When designing RPA primers, the following principles need to be followed:
[0102] 1) The length of the primer is 30-35bp. The length of the primer affects the activity of the recombinase. Too short a primer will reduce the efficiency of the recombinase. Possibility of primer secondary structure formation, which does not necessarily increase efficiency;
[0103] 2) The GC content is 40-60%. Among them, the presence of cytosine in the 3-5 nucleotides at the 5' end...
Embodiment 2
[0124] Embodiment 2RPA primer screening
[0125] In this example, three pairs of primers for TC1507 will be designed according to Twist DX The instruction manual of Liquid BasicKit was used to carry out the experiment, and a pair of primer sets that amplified better target bands were screened out.
[0126] Preparation of RPA reaction system: ddH 2 O 0.2 μL, Primer F / R 2.4 μL at a concentration of 10 μM, 25 μL of 2×ReactionBuffer, 9 μL of dNTPs at a concentration of 10 mM, 5 μL of 10×Basic E-mix, 2.5 μL of 20×Core Reaction Mix and 2.5 μL of MgOAc at a concentration of 280 mM, extracted Genomic DNA 1 μL.
[0127] The RPA reaction program is: 37°C, 20min.
[0128] The RPA results were detected by agarose gel electrophoresis, and the target fragment lengths of T6, T9, and T10 were 158bp, 159bp, and 161bp, respectively. Test results such as figure 1 As shown, T6, T9 and T10 are samples of different primers of transgenic maize TC1507 respectively, and N6, N9 and N10 are negati...
Embodiment 3
[0130]1) Use a DNA extraction kit (Shanghai Boman Biotechnology Co., Ltd.) and refer to the kit instructions to extract genomic DNA from transgenic corn TC1507.
[0131] 2) Prepare RPA reaction system
[0132] Set up a negative control (ddH 2 O) system, with the genome containing the target sequence of transgenic maize TC1507 as positive, prepare the RPA reaction system, set up a negative control to prove that the reaction system is not contaminated by other genomes, achieve the purpose of distinguishing positive samples, and verify the correctness of the measurement results.
[0133] In order to prevent false positive results in the experiment, when preparing the RPA reaction system and the control system, partition operations were performed.
[0134] When preparing the reaction system, without adding template, first prepare the outer primer F, inner primer R, 2×Reaction Buffer, dNTPs, 10×Basic E-mix and mix well, then add 20×Core Reaction Mix on the cap of the tube, Mix we...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com