A new method for simple and convenient detection of SNPs
A tomato and strip technology, applied in biochemical equipment and methods, recombinant DNA technology, and microbial determination/inspection, etc., can solve problems such as sample contamination, DNA quality, high technical level of DNA concentration, false positives, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
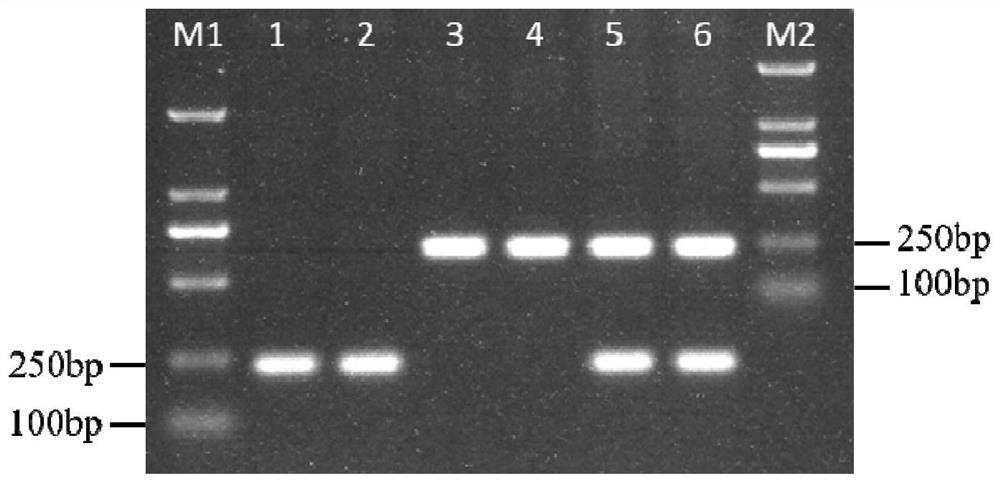
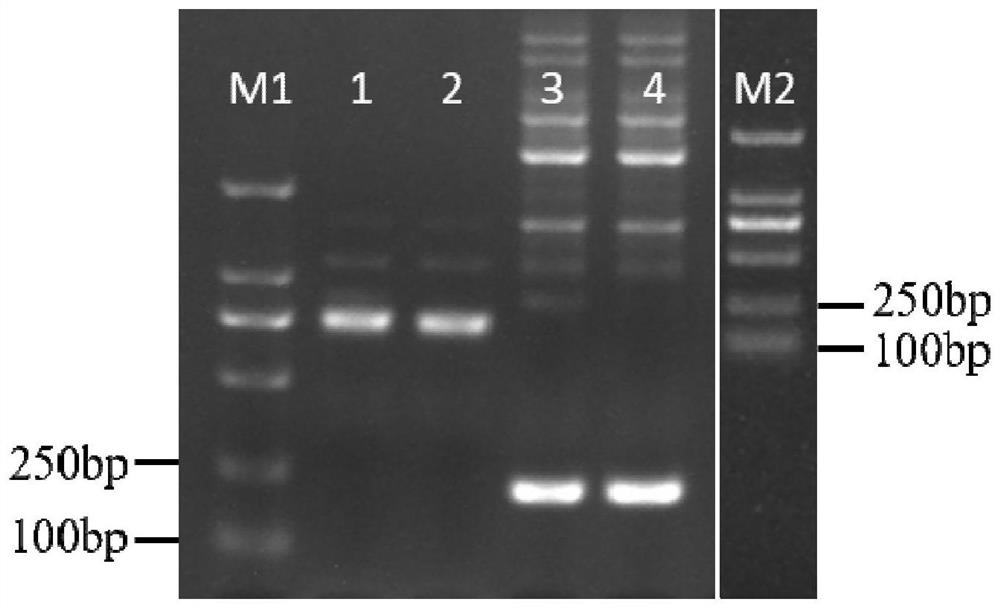
[0080] Example 1, Effect comparison of different numbers of mismatched bases in primers
[0081] Take a SNP on the first exon of the tomato Solyc03g005870.2.1 gene as an example, and the full name of the SNP is SNP144. SNP144 is an A / C polymorphism. SNP144 and its surrounding nucleotides are shown in sequence 1 of the sequence listing.
[0082] 1. Construction of recombinant plasmids
[0083] The double-stranded DNA molecule shown in Sequence 1 of the Sequence Listing (in this case, M is A) was cloned into a cloning vector to obtain recombinant plasmid A. The double-stranded DNA molecule shown in Sequence 1 of the Sequence Listing (in this case, M is C) is cloned into a cloning vector to obtain recombinant plasmid C.
[0084] 2. Design and preparation of primers
[0085] 1. Design of primers (single base mismatch primers)
[0086] The following four primers were designed: primer SNP144-C1F, primer SNP144-A1F, primer SNP144-A201F and primer SNP144-R.
[0087] Primer SNP14...
Embodiment 2
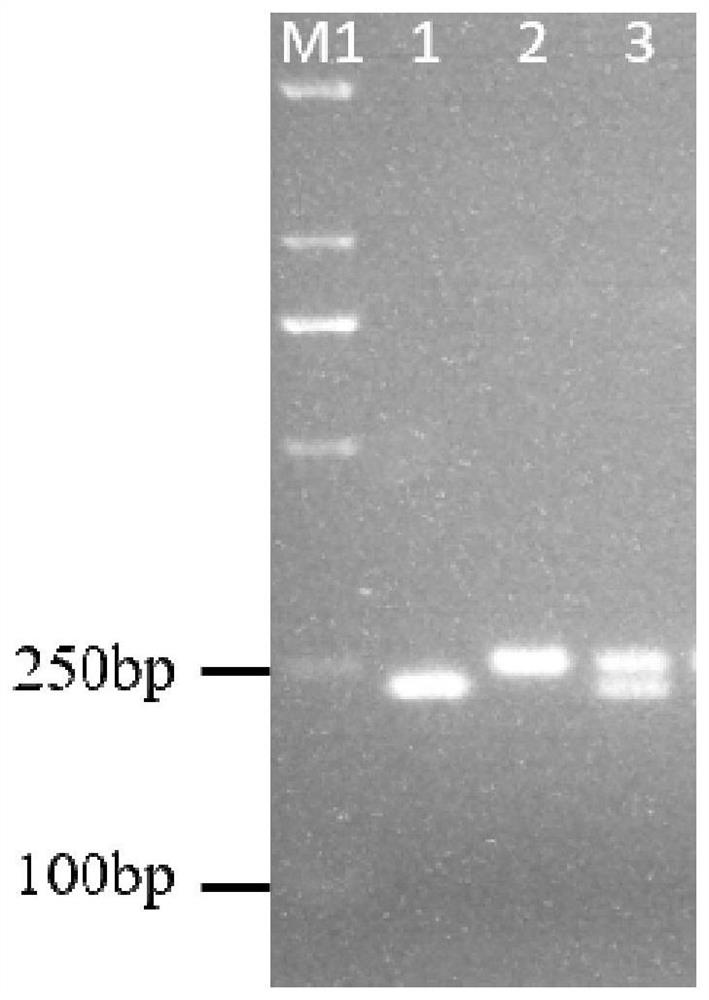
[0176] Example 2, Genotype Detection of Tomato Spotted Wilt Related SNP Marker SNP724 Site
[0177] Take a tomato spotted wilt related SNP in the tomato genome as an example, the full name of the SNP is SNP724. SNP724 is a G / A polymorphism. SNP724 and its surrounding nucleotides are shown in sequence 9 of the sequence listing. A is the resistant allele and G is the susceptible allele.
[0178] 1. Design and preparation of primers
[0179] The following four primers were designed: primer SNP724-AF, primer SNP724-GF, primer SNP724-G20F and primer SNP724-R.
[0180] Primer SNP724-AF (SEQ ID NO: 10): 5'-TCATGGACACAACTGGAGTTATgA-3';
[0181] Primer SNP724-GF (SEQ ID NO: 11): 5'-TCATGGACACAACTGGAGTTATgG-3';
[0182] Primer SNP724-G20F (SEQ ID NO: 12): 5'- GATAAAGCTTTGGAATGGAA TCATGGACACAACTGGAGTTATgG-3';
[0183] Primer SNP724-R (SEQ ID NO: 13): 5'-CTTTGTCTGGCGAAACTAGGGAACAG-3'.
[0184] Primer SNP724-AF is an upstream primer, the first nucleotide from the 3' end correspond...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com