Single cell factory capable of efficiently synthesizing L-phenylglycine as well as construction and application of single cell factory
A single-cell, factory technology, applied in the field of microorganisms, can solve the problems of affecting the transformation stability, high cost, cumbersome process, etc., and achieve the effects of low culture cost, improved transformation efficiency, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1: Preparation of Escherichia coli Competent and Transformation of Plasmid
[0035] (1) Preparation of competent Escherichia coli. Activate the monoclonal Escherichia coli in 10ml LB medium, then transfer to 37°C shaking culture to OD 600 0.35 to prepare the competent state; put the cultured bacterial solution in ice water, shake gently to cool the bacterial solution for about 10 minutes; prepare several 1.5mL centrifuge tubes that have been sterilized, and divide the bacterial solution into the tubes. The amount of bacteria in the tube is 1.2mL, put the centrifuge tube in ice; centrifuge the bacteria liquid at 8000r / min for 10-20s, let it stand in ice water for 2min, discard the supernatant, add pre-cooled 0.1M CaCl 2 400μL, gently blow the suspension, put it in ice for 15min (repeat this step 2-3 times); finally, add pre-cooled 0.1M CaCl 2 80 μL, gently pipette the suspension and place it on ice.
[0036] (2) Transformation of plasmids: Take the competent c...
Embodiment 2
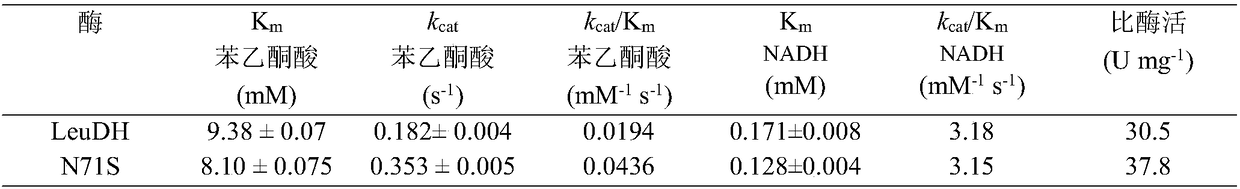
[0037] Example 2: Expression and enzyme activity determination of LeuDH mutant N71S in Escherichia coli
[0038] (1) The acquisition of mutant N71S, using the nucleotide sequence shown in SEQ ID NO.4 as a template, Fprimer (sequence shown in SEQ ID NO.5) and Rprimer (sequence shown in SEQ ID NO.6) as follows: primers, PCR is performed to obtain the recombinant gene shown in SEQ ID NO.3.
[0039] (2) The recombinant gene and pET-28a were digested with EcoR I and Xho I respectively, and after purification, they were ligated with T4 DNA ligase overnight at 16°C. The ligation product was chemically transformed into E.coli BL21 competent cells. Incubate at 37°C for 1-2 hours, apply the transformation solution to an LB plate containing kanamycin (50mg / L), extract the plasmid, and double-enzyme digest to verify the constructed recombinant plasmid, which is named pET-28a-N71S, and the sequencing work was provided by Shanghai Sangong Finish. The obtained bacterial strain containing ...
Embodiment 3
[0048] Example 3: Construction of the promoter of the FDH mutant A10C that provides the cofactor NADH and the optimized recombinant Escherichia coli of the RBS sequence
[0049] (1) The tac promoter was selected, and according to the tac promoter on the pXMJ-19 plasmid and the gene sequence of the mutant A10C, RBS sequences containing different strengths were designed (indicated in bold underline, such as SEQ ID NO: 7~SEQ ID NO: 13) PCR primers r1FDH, r2FDH, r3FDH, r4FDH, r5FDH, r6FDH and r7FDH (SEQ ID NO: 14~SEQ ID NO: 20), and the end primer pFDHRBamHI of the formate dehydrogenase gene (SEQ ID NO :twenty one).
[0050] (2) Using the existing vector pET28a-A10C carrying the FDH mutant A10C as a template, primers containing different intensities of the RBS sequence and pFDHRBamHI were used to form a primer pair, and PCR was performed to obtain multiple sequences containing RBS sequences and formate dehydrogenase The gene fragment was connected with the pXMJ-19 plasmid (the nu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com