Construction method of split-TEV-Fast system and application of split-TEV-Fast system in detecting protein interaction
A split-tev-fast and tev-fast technology, applied in the fields of biotechnology and molecular biology, can solve the problems of low reaction sensitivity, inability to accurately quantify the strength of protein interaction, and large interaction space between enzymes and substrates. The effect of sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
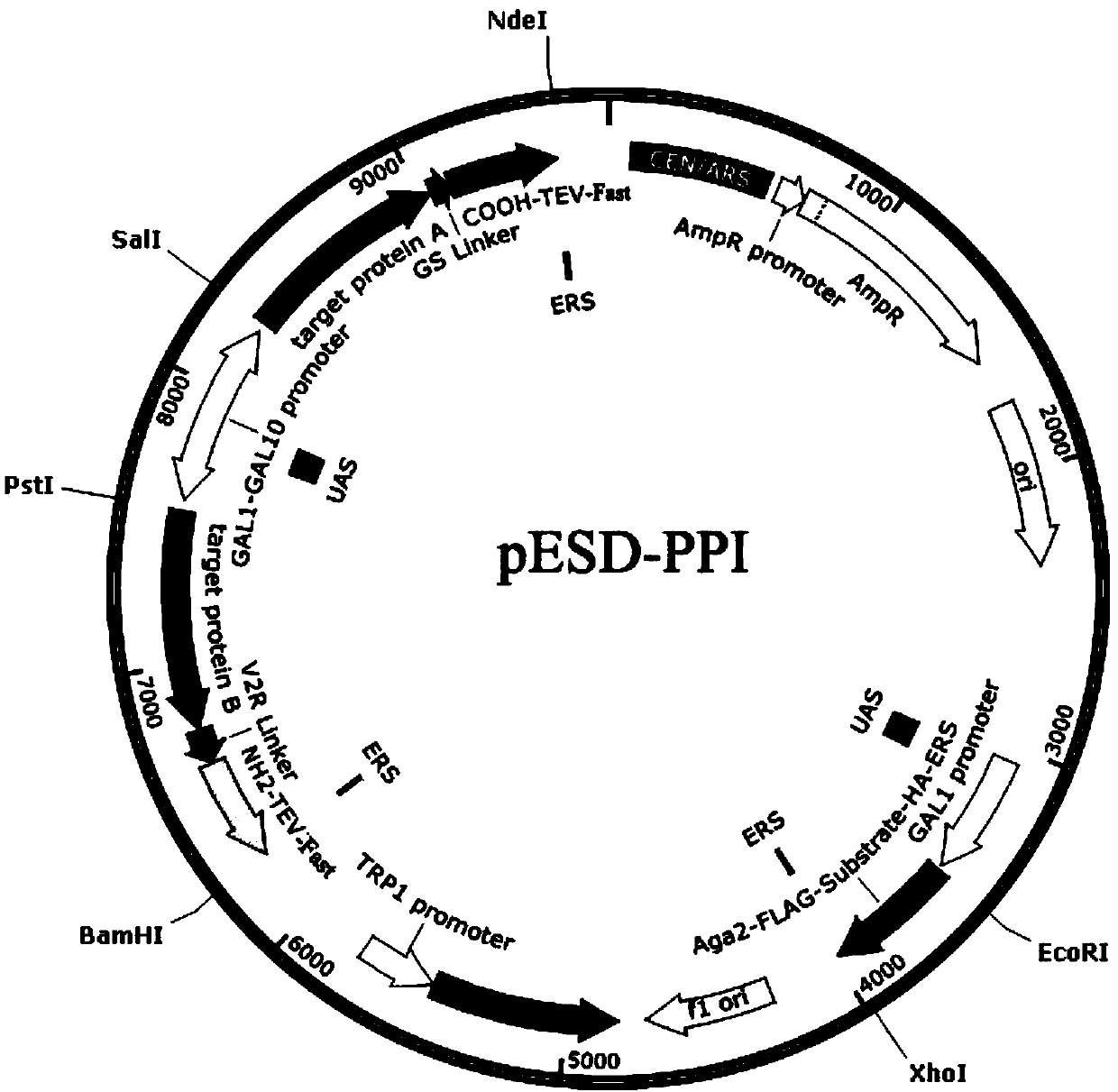
[0031] Construction process of WEHDEL recombinant vector containing retention signal peptide
[0032] (1) Firstly, the Aga2-FLAG-ENLYFQS-HA-ERS (WEHDEL) target gene was amplified by polymerase chain reaction (PCR). The reaction system is as follows: 50μL system, 10×KOD buffer, 5μL; dNTP (2.5mM) , 3μL; Primer F1 (10μM), 2μL; Primer R1 (10μM), 2μL; Pfu polymerase, 1μL; Template (containing TEV protease substrate polypeptide sequence), 0.5μL (20ng / μL); add double distilled water to 50μL .
[0033] PCR amplification conditions: 95℃, 5min; 95℃, 30s, 57℃, 30s, 72℃, 30s, 25 cycles; 72℃, 5min; 4℃, ∞.
[0034] The primer design is as follows:
[0035] F1 is SEQ ID NO.1
[0036] R1 is SEQ ID NO. 2
[0037] (2) The target fragments were recovered by agarose gel, which were digested with XhoI and EcoRI at 37°C for 6 hours, and then recovered with agarose gel. Double digestion system: target fragment DNA, 30μL; Buffer Cutsmart, 5μL; XhoI, EcoRI, 2μL each; add double distilled water to 50μL.
[0038...
Embodiment 2
[0061] Induced expression of recombinant vector containing Split-TEV-Fast system
[0062] The reagents used to transform the final vector into Saccharomyces cerevisiae EBY200 transformation are: EBY200 competent, 20μL; recombinant plasmid, 1.5μL; Single strand carrier DNA, 25μL; 1M LiAC, 36μL; PEG4000 (50%W / V), 240μL; mix After the system, 30 ℃ water bath for 30 min, transfer to 42 ℃ heat shock for 20 min, centrifugation to collect the bacteria, add 1mL YPD liquid medium to 30 ℃ shaker culture for 1.5 hours, wash the cells with ultrapure water once, then apply to YNB-CAA- Glucose solid medium, 30℃ incubator for 2-3 days. The composition of YNB-CAA-Glucose solid medium is 20g / L glucose, 6.7g / L YNB, 5.4g / L Na 2 HPO 4 ,8.6g / L NaH 2 PO 4 ·H 2 O, 5g / L casamino acids, pH7.4, 15g / L agar.
[0063] Inoculate a single colony in the liquid medium YNB-CAA-Glucose, culture it in a shaker at 30°C, 230rpm until OD600=3, transfer to YNB-CAA-Galactose liquid medium, cultivate it in a shaker at 30°...
Embodiment 3
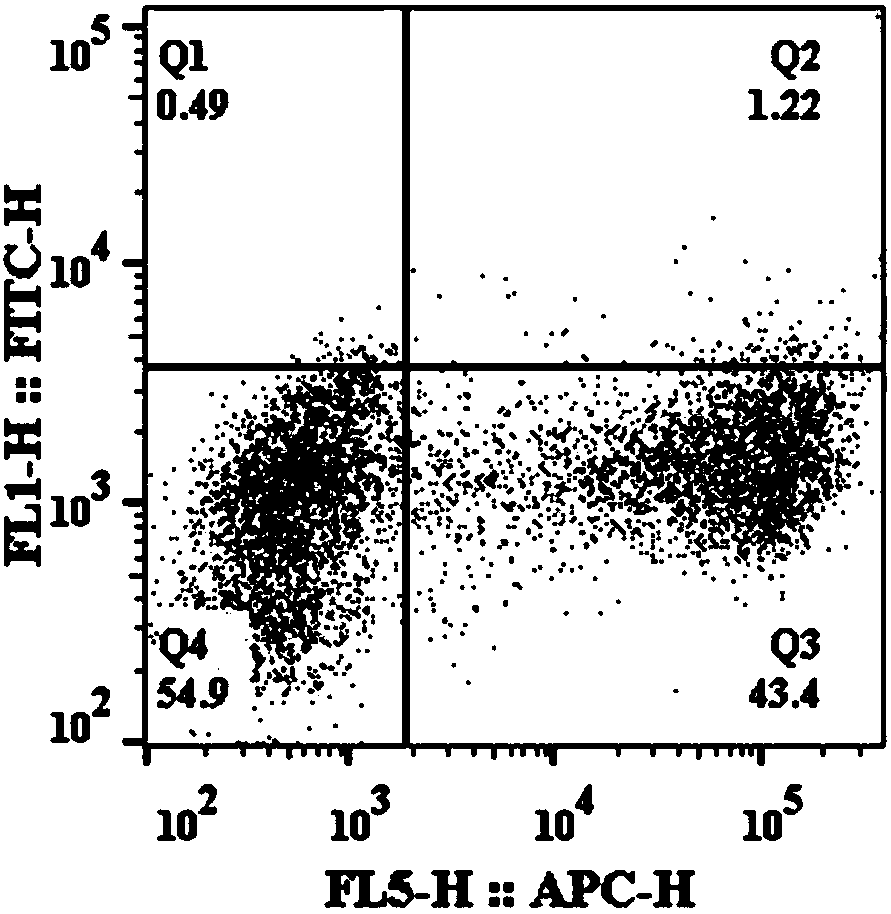
[0065] Use flow cytometry to detect the results of the Split-TEV-Fast system to detect protein interactions
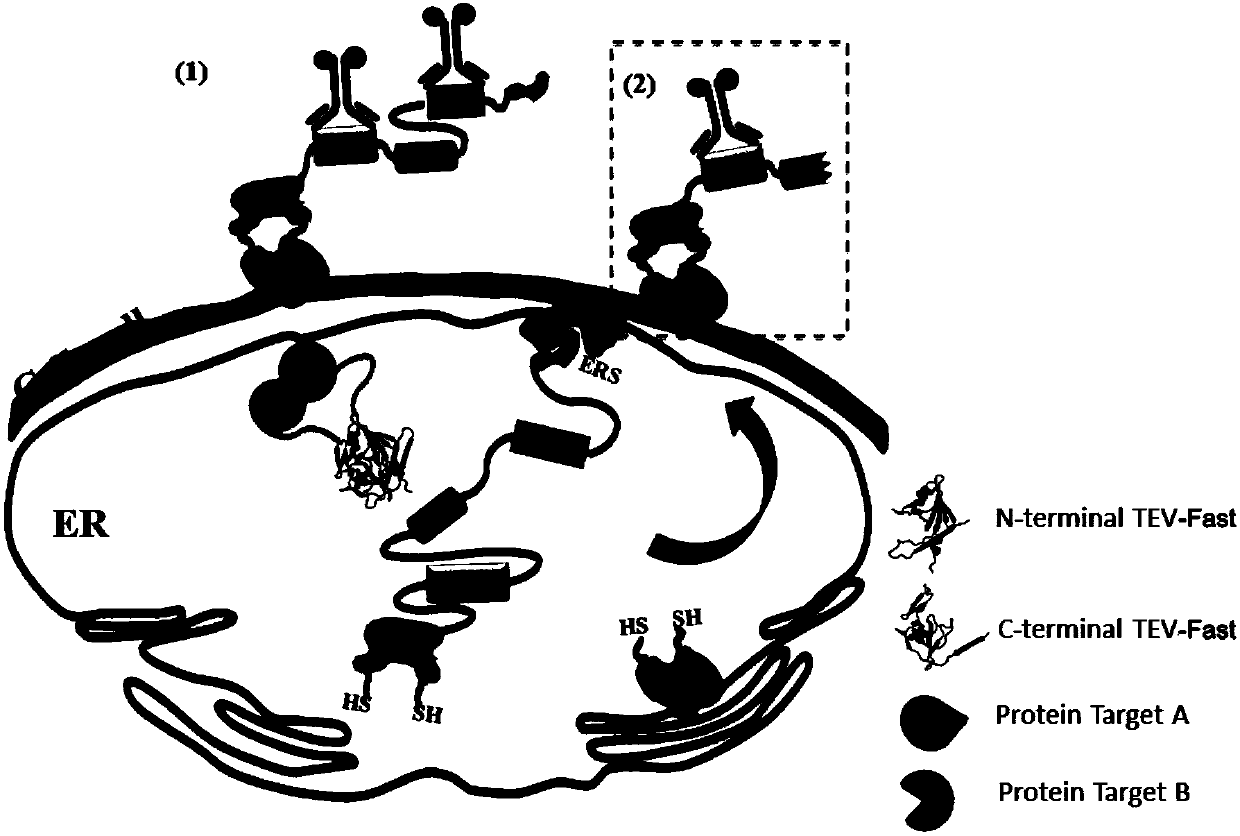
[0066] A complex composed of Yael, Lto1, and Rli1 was selected to verify the Split-TEV-Fast system, where Lto1 and Yael interact with each other, but not with Rli1. Therefore, Yae1 and Lto1 are set as the experimental group, and Rli1 and Lto1 are the negative control groups. Due to the interaction between Yae1 and Lto1, the split-TEV-Fast -NH2 end and -COOH end are reconstituted to form a complete and active TEV- Fast is to cleave the substrate, and the yeast surface displays Aga2-FLAG-ENLYFQ, and then the single fluorescent signal iFluor 647 is detected by the Anti-FLAG-iFluor 647 fluorescent antibody label. When the target protein is Rli1 and Lto1, if the two do not interact, a complete and active TEV-Fast protease cannot be formed, and -ENLYFQS cannot be cleaved. The yeast surface displays Aga2-FLAG-ENLYFQS-HA Then, the dual fluorescence signal (iFluor 647 and FITC) w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com