Host removal extraction and database building method for sputum microorganism metagenome
A technology of metagenomics and extraction methods, which is applied in the field of host-free extraction and library construction of sputum microbial metagenomics, which can solve the problems of low initial quantity, inability to meet multiple library types and multiple sequencing platforms at the same time, and achieve compatibility Good performance, beneficial to the effect of quantitative stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
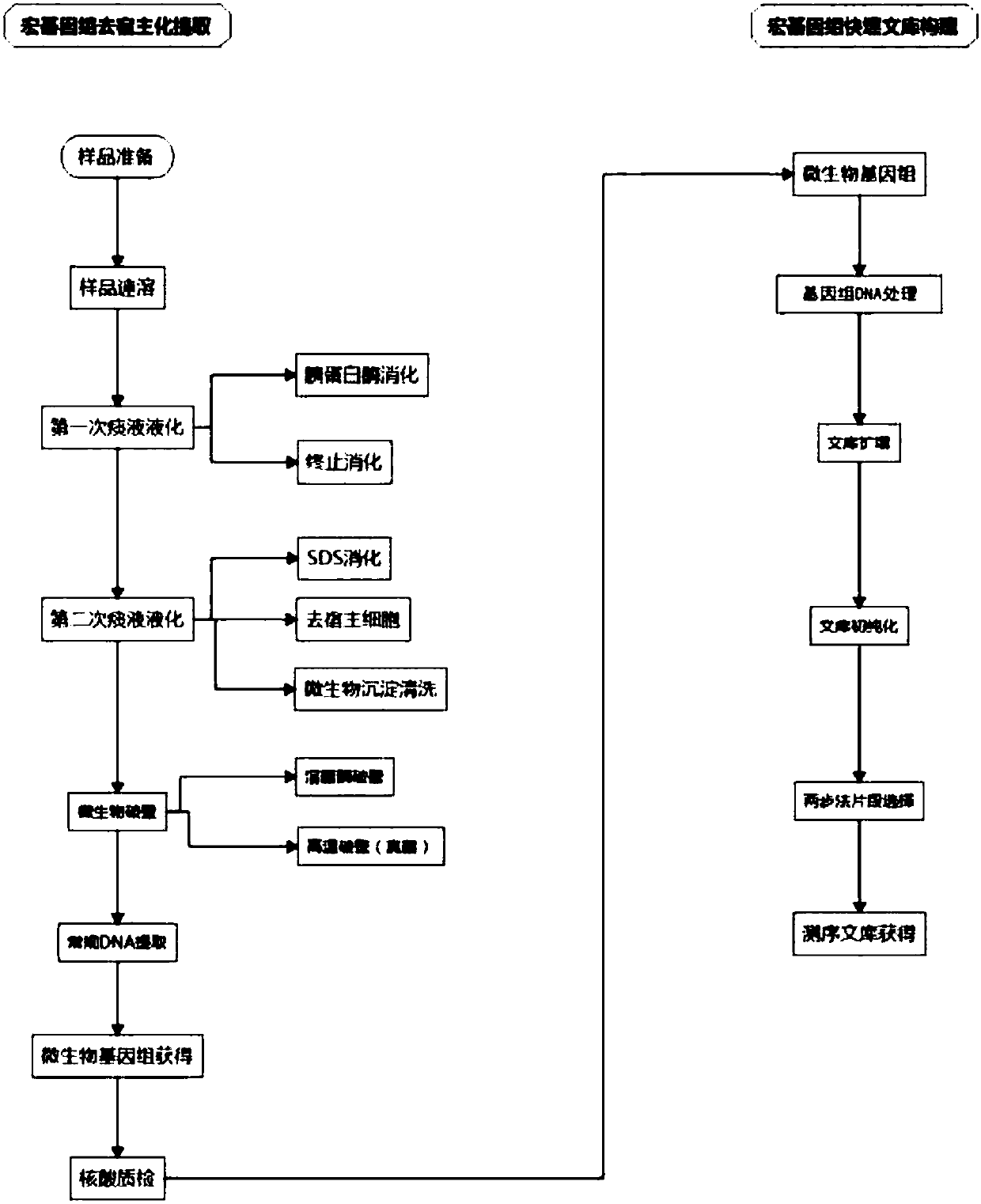
[0050] A method for extracting host-free microbial metagenomic groups from sputum, specifically comprising the following steps:
[0051] 1. Sample pretreatment
[0052] (1) Sample preparation: The sample materials are about 60 cases of sputum and airway lavage fluid from a group of patients with severe pneumonia provided by Nanfang Hospital of Southern Medical University. About 2 mL of sputum sample was loaded into a 5 cm high cylindrical sputum collection tube, concentrated at the bottom, and stored at -80 °C.
[0053] (2) Sample instant dissolution: place the bottom of the sputum collection tube in a 37°C water bath and shake continuously to make the sample change from solid to dissolved liquid within 2 minutes. If the sample is in a frozen state, instant dissolution is required, and if it is at 4°C or a fresh sample, this step can be omitted. Instant thawing allows the sample to be thawed in a short time without destroying the sample itself and its cellular integrity.
...
Embodiment 2
[0105] A method for constructing a sputum microbial metagenomic library, specifically comprising the following steps:
[0106] 1. Sample genomic DNA extraction
[0107] Extract with reference to the method of embodiment 1.
[0108] 2. Genomic DNA processing
[0109] (1) Add 10 µL of reaction buffer and 5 µL of gDNA (accurately quantified 1 ng of total QB) to mix, then add 5 µL of transposase mixture (ATM), and mix well;
[0110] (2) The mixture was reacted on a PCR machine for 6 minutes at 55°C to fragment the genome into a length of 230 bp, and then 5 µL of termination reaction buffer (NT) was added to terminate the enzyme reaction.
[0111] 3. Library amplification
[0112] (1) Add a combination of index primers to the processed genomic DNA;
[0113] (2) Add 15µL PCR Enzyme Master Mix, mix and centrifuge;
[0114] (3) React the mixture on a PCR instrument. The PCR amplification conditions are: 72°C for 3 minutes; 95°C for 30s; 95°C for 10s, 55°C for 30s, 72°C for 30s, 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com