A method for dehosting extraction and library construction of sputum microbial metagenomics
A technology of metagenomics and extraction methods, which is applied in the field of host-free extraction and library construction of sputum microbial metagenomics. Cost, good prospects for promotion and application, and good compatibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
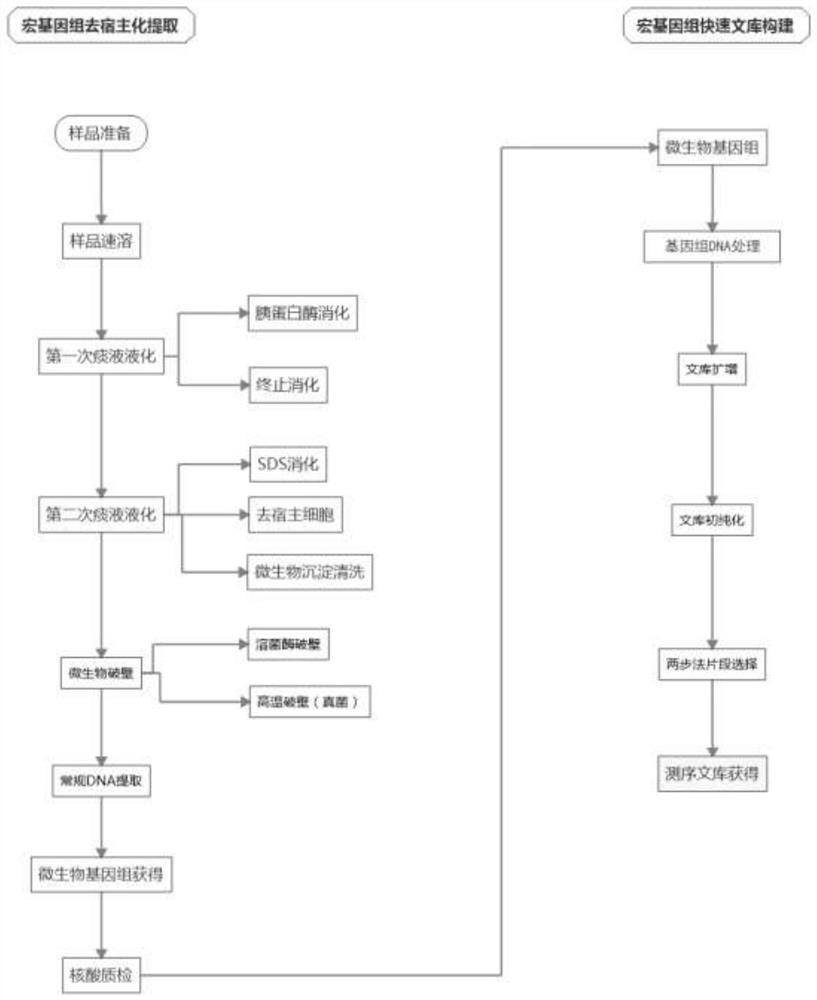
[0050] A sputum microorganism metagenome dehosting extraction method, specifically comprising the following steps:
[0051] 1. Sample pretreatment
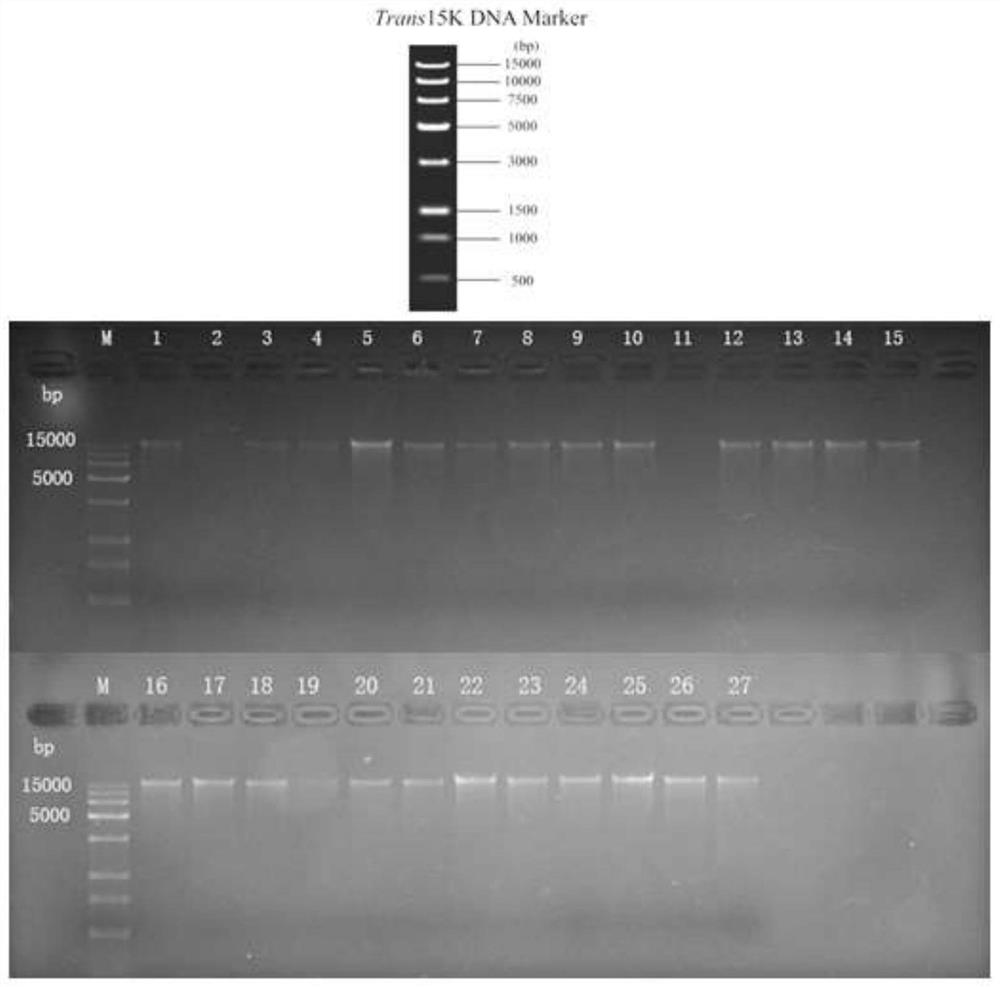
[0052] (1) Sample preparation: The sample materials were about 60 cases of sputum and airway lavage fluid from a group of severe pneumonia patients provided by the Southern Hospital of Southern Medical University. About 2 mL of sputum sample was loaded into a 5 cm high cylindrical sputum collection tube, collected at the bottom, and stored at -80°C.
[0053] (2) Instant dissolving of the sample: place the bottom of the sputum collection tube in a 37°C water bath and shake continuously to make the sample change from a solid state to a dissolved liquid state within 2 minutes. If the sample is in a frozen state, it needs to be instant-dissolved. If it is a 4°C or fresh sample, this step can be omitted. The instant solution allows the sample to be thawed in a short time without destroying the sample itself and its cellular integrity...
Embodiment 2
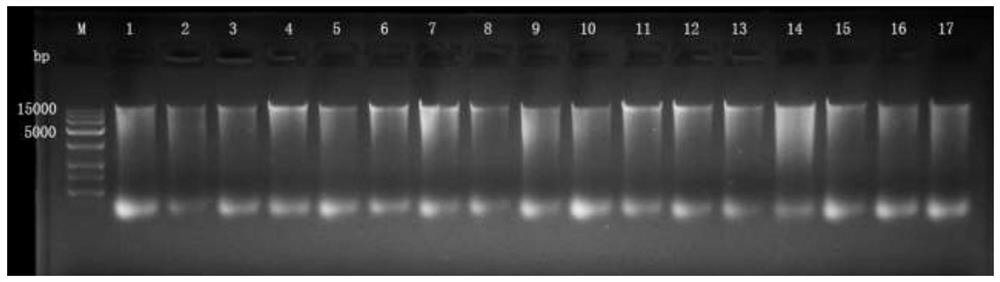
[0105] A sputum microbial metagenomics method for building a library, specifically comprising the following steps:
[0106] 1. Sample genomic DNA extraction
[0107] Extract according to the method of Example 1.
[0108] 2. Genomic DNA processing
[0109] (1) Add 10µL of reaction buffer and 5µL of gDNA (total QB accurately quantified 1ng) and mix, then add 5µL of transposase mixture (ATM), and mix well;
[0110] (2) The mixture was reacted on a PCR machine at 55°C for 6 minutes, the genome was fragmented to a length of 230bp, and then 5µL of termination buffer (NT) was added to terminate the enzymatic reaction.
[0111] 3. Library amplification
[0112] (1) adding a combination of index primers to the treated genomic DNA;
[0113] (2) Add 15µL PCR amplification enzyme master mix, and mix and centrifuge;
[0114] (3) The mixture was reacted on a PCR instrument, and the PCR amplification conditions were: 72°C for 3 min; 95°C for 30s; 95°C for 10s, 55°C for 30s, 72°C for 30s...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com