Biosensor capable of responding to 3-dehydroshikimic acid and application of biosensor
A biosensor, dehydroshikimic acid technology, applied in DNA/RNA fragments, recombinant DNA technology, bacteria, etc., can solve the problem of unreported positive correlation between cusR and 3-dehydroshikimic acid, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Example 1: Transcriptome analysis mining 3-dehydroshikimic acid response function regulatory genes
[0065] Take the 3-dehydroshikimic acid-producing strain E.coli WJ060 as the experimental strain and the non-3-dehydroshikimic acid-producing strain E.coli ATCC 8739 as the control strain, pick a single colony and inoculate the LB medium, shake the flask at 37°C Fermentation 24h, bacterial strain E.coli WJ060 selects the fermentation medium (WJ060-12h and WJ060-20h) of two sampling points of fermentation culture 12h and 20h, bacterial strain E.coli ATCC 8739 selects the fermentation medium of fermentation culture 12h sampling point (8739 -12h). The fermentation broth was centrifuged at 4°C and 3200rpm for 5 minutes, and the bacterial cells were quickly frozen in liquid nitrogen and sent for transcriptome sequencing analysis.
[0066] In order to mine 3-dehydroshikimate-responsive functional genes, the bioinformatics software Bowtie2 was used to compare the three transcri...
Embodiment 2
[0072] Example 2: Real-time PCR verification of transcriptional regulatory genes responding to 3-dehydroshikimic acid
[0073] 1) Real-time PCR verification of transcriptional regulatory genes in response to endogenous production of 3-dehydroshikimate
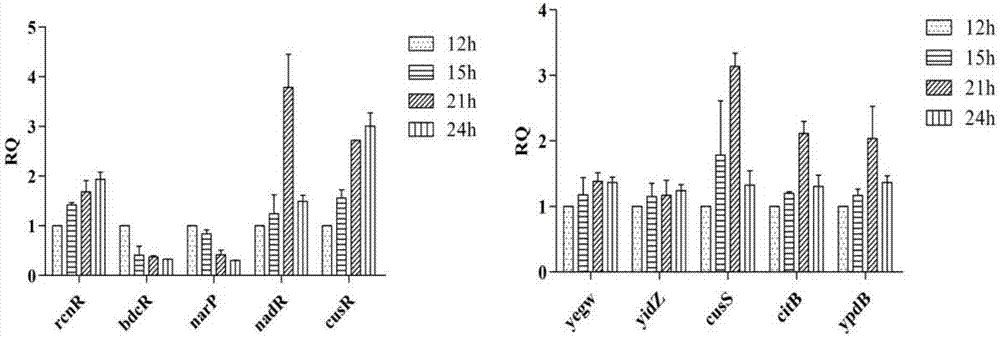
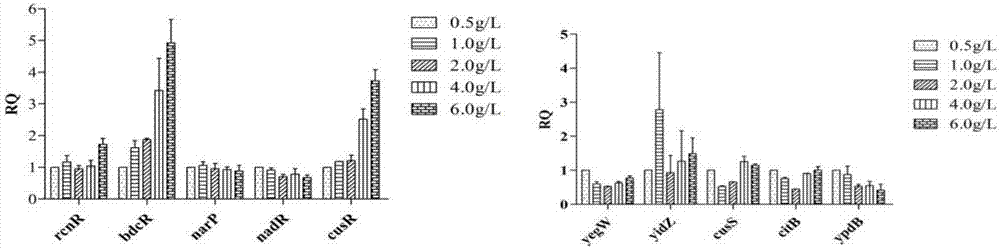
[0074] The 3-dehydroshikimate-producing strain E.coli WJ060 was used as the experimental strain, and a single colony was picked and cultured in LB medium at 37°C overnight. Then transfer the NBS medium with 2% inoculum of 2% overnight culture solution, shake the flask for fermentation and culture at 37°C, take samples at four time points of 12h, 15h, 21h, and 24h, and control the 10 cells according to the real-time PCR operation process. Genes were verified by real-time PCR, and the responses of 10 transcriptional regulatory genes to the continuous accumulation of intracellular 3-dehydroshikimic acid product concentration during fermentation were analyzed.
[0075] The result is as figure 2 , at the four fermentation time po...
Embodiment 3
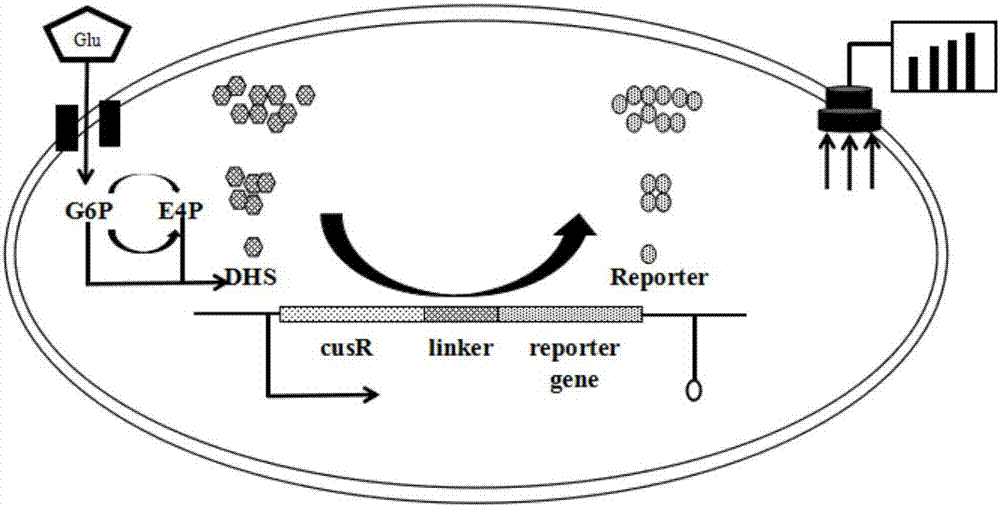
[0081] Example 3: Construction of 3-dehydroshikimic acid biosensor
[0082] In order to construct a 3-dehydroshikimate-sensing biosensor, it is first necessary to obtain a DNA fragment of the 3-dehydroshikimate-responsive functional fusion element, which includes the regulatory gene cusR, the fluorescent reporter gene gfp, and the linker sequence L (Linker) part. Among them, the intermediate connection sequence L connecting the functional response regulatory gene cusR and the gfp reporter gene is designed as a short peptide of 15 amino acids, and the coding sequence of the gfp gene without the start codon and the coding sequence of the positively related gene cusR without the stop codon The coding sequence of L is 5'-ggtggtggtggttctggtggtggtggatccggtggcggtggttct-3.
[0083] In order to obtain the cusR gene fragment of the regulatory gene, this experiment used the genome of the strain E.coli ATCC 8739 as a template, and used the upstream primer sequence: 5'-ccggaattcggctgagtga...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com