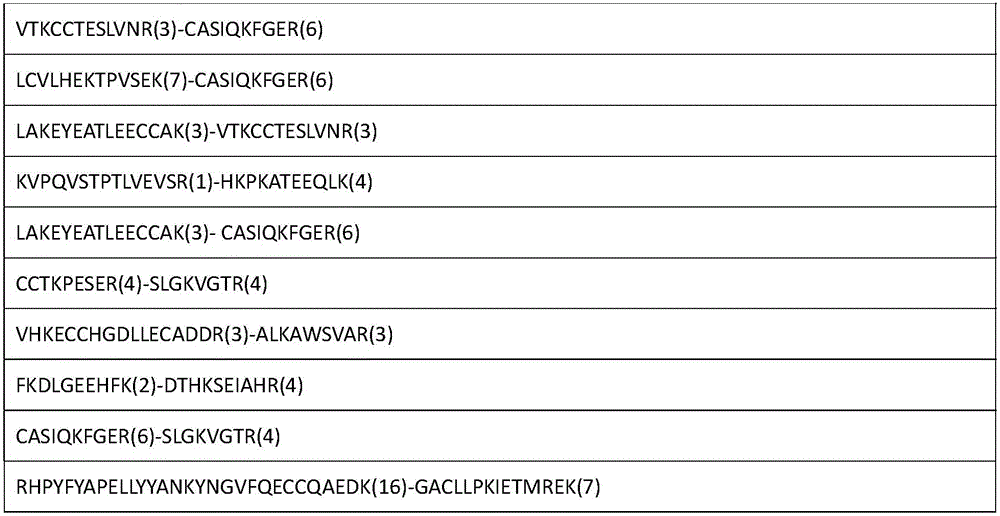
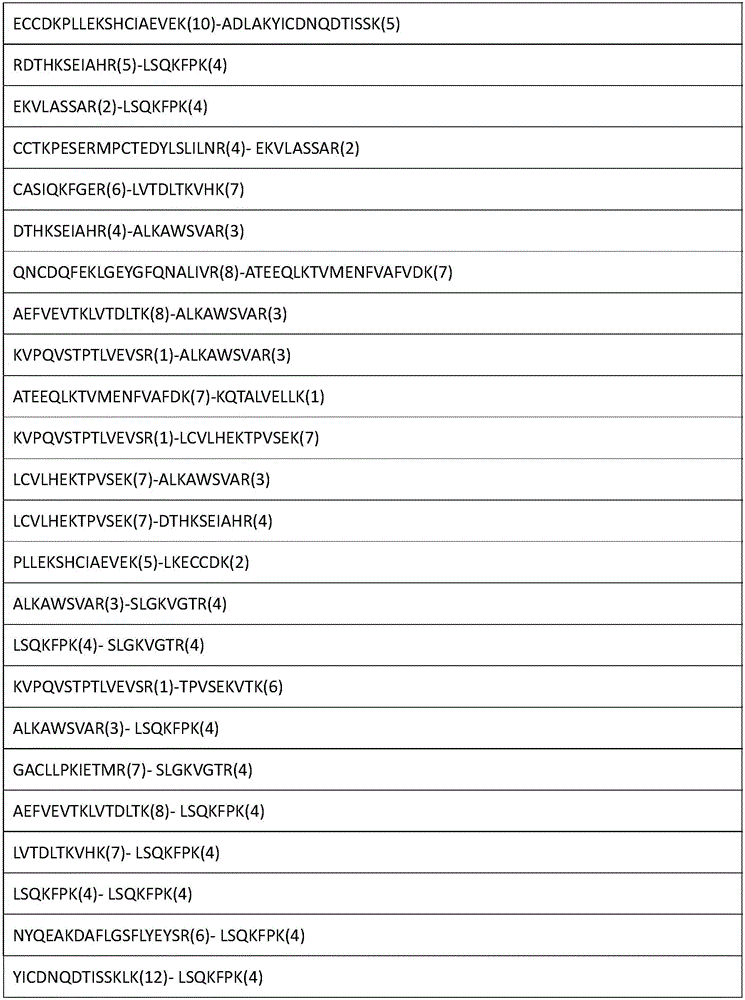
Analytical method for researching protein structure or protein-protein interaction
A protein structure and protein technology, which is applied in the analysis field of studying protein structure or protein interaction, can solve the problems of inability to realize the analysis of the whole proteome crosslinking information, difficult to realize the identification of peptide crosslinking, and many mismatches in the identification results. Identify analysis time reduction, scale reduction, and effect of scale reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] 1. Cross-linking reaction of protein samples
[0027] Dissolve 10 μg of bovine serum albumin sample (BSA) in 20 mM 4-hydroxyethylpiperazineethanesulfonic acid buffered saline solution (HEPES) at pH 7.4 to a final protein concentration of 1 mg / mL and use dimethyl sulfoxide (DMSO) Dissuccinimidyl tartrate (DST) was prepared at a concentration of 25 mM, the cross-linking agent was added to the BSA solution to make the final concentration 1 mM, and reacted at room temperature for 1 h.
[0028] 2. Removal of excess cross-linking agent
[0029] Ammonium bicarbonate solution (ABC) was added to the reaction solution in step 1 to make the final concentration 50 mM to terminate the cross-linking reaction.
[0030] 3. Dissolution, denaturation and reduction of protein samples
[0031]Add urea and DTT to the cross-linked BSA solution in step 2, so that the final concentration of urea in the solution is 8M, and the final concentration of DTT is 10mM, and react in a water bath at 3...
Embodiment 2
[0050] 1. Cross-linking reaction of BSA protein sample; removal of excess cross-linking agent; steps of dissolution, denaturation and reduction, alkylation and enzymatic hydrolysis of protein sample are the same as in Example 1.
[0051] 2. Derivatization, fractionation and mass spectrometry analysis of cross-linked peptides
[0052] An aliquot of the lyophilized BSA cross-linked hydrolyzate from step 1 was taken and redissolved with 100 mM triethylamine-carbonic acid buffer (TEAB), pH 8. After dimethylation reaction, desalting, lyophilization, samples were fractionated using cation exchange separation, desalting, lyophilizing and reconstitution using 0.1% formic acid solution for mass spectrometry analysis.
[0053] 3. Fragmentation and enrichment of cross-linked peptides, removal of non-specifically adsorbed peptides, and release of peptides are the same as in Example 1.
[0054] 4. Fractionation of enriched cross-linked peptides
[0055] The enriched cross-linked peptides...
Embodiment 3
[0063] 1. Cross-linking reaction of protein samples
[0064] 10 μg of rabbit creatine kinase protein sample (CK) was dissolved in 50 mM phosphate-buffered saline (PBS) with a pH of 7.4, and the final protein concentration was 1 mg / mL. DST was prepared at a concentration of 25 mM using dimethylformamide (DMF). The cross-linking agent was added to the CK solution so that the final concentration was 1 mM, and reacted at room temperature for 1 h.
[0065] 2. Removal of excess cross-linking agent
[0066] Tris buffer solution (Tris) was added to the reaction solution in step 1 to make the final concentration 50 mM to terminate the cross-linking reaction.
[0067] 3. Dissolution, denaturation and reduction of protein samples
[0068] Add urea and DTT to the cross-linked CK solution in step 2, so that the final concentration of urea in the solution is 8M, and the final concentration of DTT is 10mM, and react in a water bath at 37°C for 30min.
[0069] 4. Alkylation and enzymatic h...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com