Method for identifying sika deer, red deer or other hybridized deer on basis of COI and SRY sequences
A sika deer, SRY gene technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve problems such as inability to identify hybrid deer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Example 1. SNP sites for identifying sika deer, red deer and their hybrid deer and primers for amplifying them
[0078] The SRY gene is a sex-determining gene in mammals, and the location, size, and structure of the SRY gene vary among different species. Therefore, using COI and SRY genes to establish molecular identification of sika deer, red deer and their hybrid deer has become a new research direction.
[0079] 1. Discovery and application of SNP loci for identifying sika deer, red deer and their hybrid deer
[0080] According to the COI and SRY gene sequences of the tested samples obtained by sequencing, the COI and SRY gene sequences of sika deer, red deer, reindeer, sambar and other species were downloaded from the NCBI database, and homologous alignment was performed using ClustulX 2.1 software. Then analyze its specific SNP sites, and screen suitable identification sites from the specific SNP sites.
[0081] After comparison, the 391st position of the COI gen...
Embodiment 2
[0098] Example 2, Establishment of Molecular Identification Method for Antler Samples of Sika Deer, Red Deer and Their Hybrid Deer
[0099] 1. Obtaining the fragment to be detected
[0100] 1) Extraction and purification of genomic DNA
[0101] The velvet antler medicinal sample was taken, and after surface disinfection with 75% ethanol, 20 mg sample powder was drilled with a bone drill, and the decoction pieces were directly crushed. The total DNA of velvet antler was extracted by using TIANDZ column bone DNAout, which was used as a template for the identification of velvet antler DNA.
[0102] 2) Universal primers amplify the COI gene fragment to be detected and the SRY gene fragment to be detected
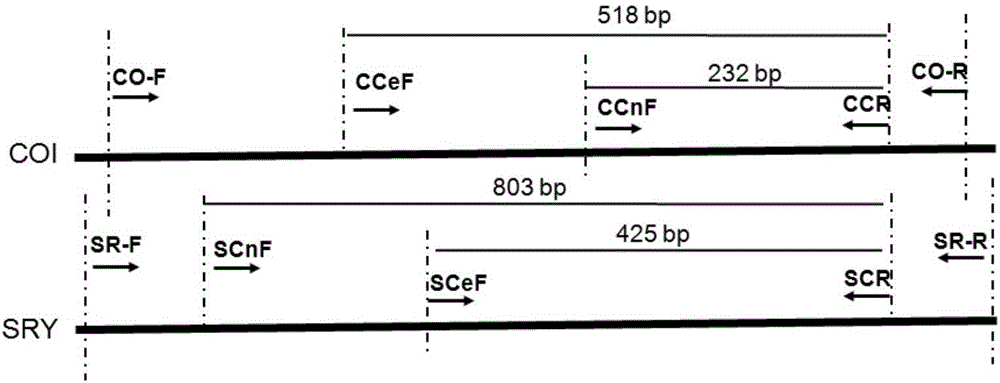
[0103] Using the total DNA obtained in the above 1) as a template, use the universal primers CO-F and CO-R (see Table 2) to amplify the COI gene sequence. The PCR reaction program is: 95°C, 5min; 95°C, 30s, 45°C, 1.5min, 72°C, 1.5min, 5 cycles; 95°C, 1min, 50°C, 1.5min, 72°C, ...
Embodiment 3
[0125] Embodiment 3, multiplex PCR distinguishes tested sika deer, red deer and its hybrid deer samples
[0126] The tested samples were 8 batches of known sika deer antler samples, 16 batches of known red deer antler samples and 3 batches of horse-flower hybrid antler samples collected in the heavy circulation market.
[0127] The method of Example 2 was used to carry out multiplex PCR detection on the test samples.
[0128] If the multiplex PCR system for amplifying COI amplifies a specific fragment of 232bp, and the multiplex PCR system for amplifying SRY amplifies a specific fragment of 803bp, then the velvet sample to be tested comes from or the candidate is from sika deer; otherwise, the velvet to be tested The sample does not originate or the candidate does not originate from sika deer;
[0129] If the multiplex PCR system for amplifying COI amplifies a specific fragment of 518bp, and the multiplex PCR system for amplifying SRY amplifies a specific fragment of 425bp, t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com