Application of FXR antagonist in preparing anti-HBV medicines
An antagonist and antiviral drug technology, applied in the field of new drug research and development, can solve problems such as drug resistance and HBV replication rebound, and achieve the effects of reducing economic burden, improving liver function, and promoting HBV clearance.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
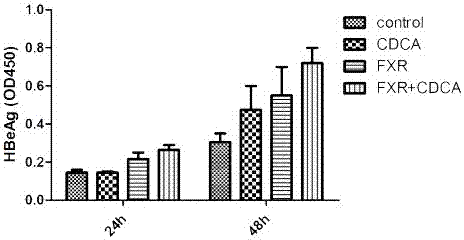
[0019] CDCA (chenodeoxycholic acid) promotes HBV expression through FXR in cell models:
[0020] Experimental materials: HepG2, pHBV1.3, pFXR, pGFP, HBV detection kit (Shanghai Kehua), CDCA (Sigma).
[0021] Experimental method: In HepG2 cells, co-transfect pHBV1.3, pFXR plasmid or pGFP control plasmid, 24 hours after transfection, observe the transfection efficiency with a fluorescence microscope, then add CDCA to stimulate for 24 hours and 48 hours, collect the supernatant, ELISA detection of HBeAg expression. Specifically include the following steps:
[0022] ①Equilibrate the kit at room temperature for 30 minutes;
[0023] ② Take the cell supernatant, add 50 μl to each well, and set 2 wells each for the negative control and positive control, and 1 well for the blank control;
[0024] ③ Add 50 μl of enzyme conjugate to each well (not add to the blank well), mix well, and seal the plate;
[0025] ④ Incubate in a 37°C incubator for 1 hour;
[0026] ⑤ Pour off the liquid,...
Embodiment 2
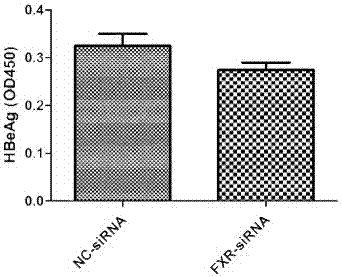
[0031] In cell models, interfering with FXR expression inhibited HBeAg expression instead:
[0032] Experimental materials: HepG2, pHBV1.3, siRNA FXR (Guangzhou Ruibo), CDCA (Sigma), HBV detection kit (Shanghai Kehua).
[0033] Experimental method: In HepG2 cells, pHBV1.3 and siRNA FXR were co-transfected, 24 hours after transfection, then stimulated with CDCA for 48 hours, the supernatant was collected, and the expression of HBeAg was detected by ELISA.
[0034] The experimental result of embodiment 2 is as figure 2 , in the HepG2 cell line, pHBV1.3 and FXR siRNA were co-transfected, CDCA was added 24 hours after transfection, and the supernatant was taken 48 hours after the addition of the drug, and the expression of HBeAg was detected by ELISA. Interference with FXR expression inhibited HBeAg expression instead (P< 0.05).
Embodiment 3
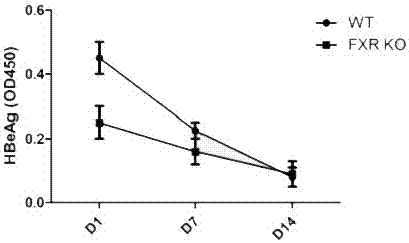
[0036] FXR Regulates HBV Expression in HBV Mouse Model
[0037] Experimental material: FXR - / - Mice (purchased from Jackson Laboratory, USA), C57BL / 6J mice (Beijing Weitong Lihua), pHBV1.3, HBV detection kit (Shanghai Kehua).
[0038] experimental method:
[0039] The HBV replication model was established by injecting HBV clones into the high-pressure tail vein: the tail was sterilized with 75% alcohol, and then irradiated with infrared light to cause varicose veins in the mouse tail, which were clearly visible. Insert the needle parallel to the tail vein, inject 8-10% of the liquid, and finish the injection within 5-8 seconds. Blood was collected on the 1st, 7th, and 14th day after the injection, and 150 μl of 1% pentobarbital sodium was injected intraperitoneally. Incubate at 37°C for 1 hour, then centrifuge at 8000rpm for 10 minutes, and store the separated serum at -80°C. ELISA detects serum HBeAg level, and the test results are as follows: image 3 , FXR expression l...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com