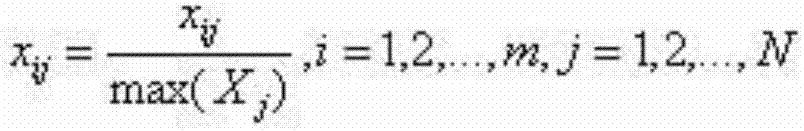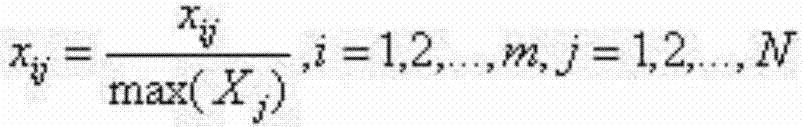Metagenome fragment attribute reduction and classification method based on neighbourhood rough set
A technology of neighborhood rough sets and metagenomics, which is applied in the field of attribute reduction and classification of metagenomic fragments based on neighborhood rough sets, can solve problems such as inability to obtain genetic information, improve classification accuracy, keep classification accuracy unchanged, The effect of reducing the amount of data
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
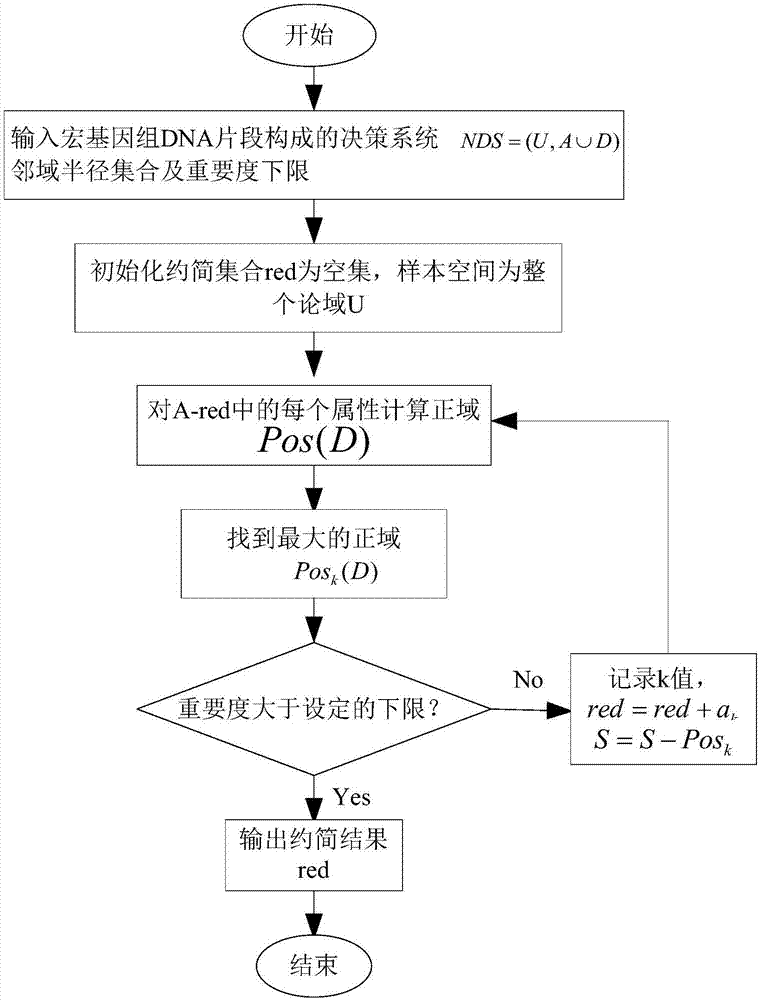
[0017] The present invention will be further described in detail below in conjunction with the accompanying drawings and specific embodiments.
[0018] A. Download the whole genome sequences of 10 kinds of microorganisms from the US National Center for Biotechnology (NCBI: US National Center for Biotechnology Information). The downloaded microbial genome sequences are all linear sequences composed of four nucleotides A, T, G, and C. Sequence; 100 non-overlapping DNA fragments with a length of 1000bp (base points) were randomly cut out from each bacterium; the four nucleotides A, T, G, and C in the metagenomic fragment were converted into numbers 0, 1, 2, 3; The meaning of k-mer frequency is the number of occurrences of K nucleotides in the gene sequence. Studies have shown that the K-mer frequency distribution of DNA fragments derived from the same species is similar, and the K-mer frequency of DNA fragments derived from different species The distribution is different, so the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com