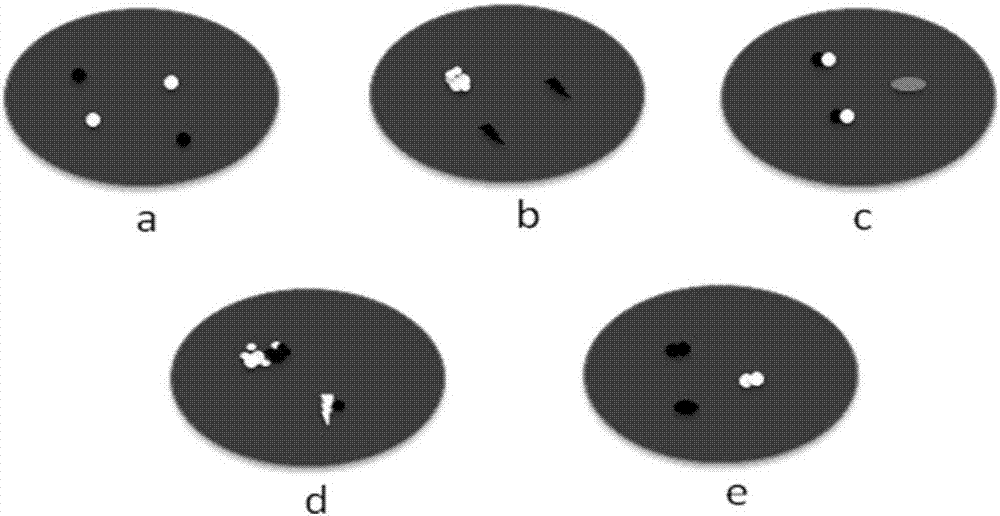
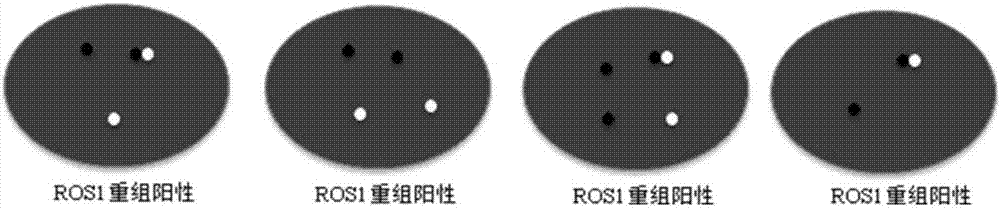
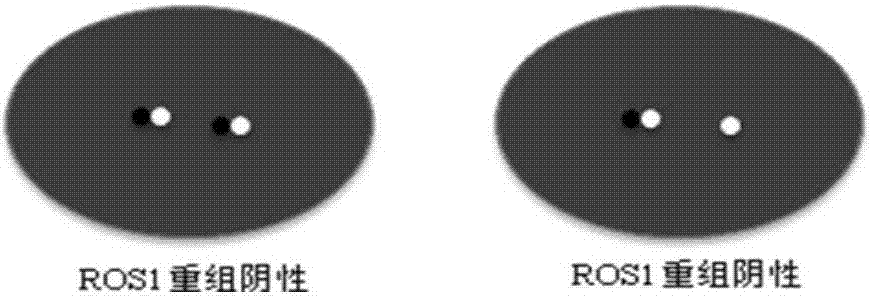
Kit and detection method for abnormity of ROS1 and C-met genes
A technology of abnormal detection and kit, applied in the field of molecular biology, which can solve the problems of large probe length, increased detection and waiting time for results, etc., to improve detection sensitivity, shorten the time required for sufficient hybridization, and ensure the specificity of results and the effect of sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 A ROS1, C-met Gene Abnormal Detection Kit
[0046] The ROS1, C-met gene abnormality detection kit described in this embodiment mainly includes:
[0047] 1. Design the amplification primers
[0048] The first and second sets of amplification primers were designed for the upstream (5' end) and downstream (3' end) of the breakpoint of the ROS1 gene respectively, wherein the first group was for the amplification primer upstream (5' end) of the breakpoint of the ROS1 gene. Amplifying primers, the second group is for amplification primers downstream of the breakpoint (3' end), and the corresponding amplification products constitute the first group of probe libraries and the second group of probe libraries; respectively for the C-met gene and Design the third and fourth sets of amplification primers for the centromere region of human chromosome 7, in which the third set is the amplification primer for the C-met gene, and the fourth set is for the amplification of th...
Embodiment 2
[0079] Example 2 Detection of clinical samples from patients with lung cancer using the ROS1, C-met gene abnormality detection kit in Example 1
[0080] In this example, the detection of ROS1 and C-met gene abnormalities in circulating tumor cells of lung cancer patients was taken as an example. The ROS1 and C-met gene abnormality detection kits in Example 1 were used to examine the peripheral blood samples of 20 lung cancer patients. Proceed as follows:
[0081] 1. Sample pretreatment
[0082] 1. Enrichment of circulating tumor cells: The peripheral blood sample is lysed with commercially available red blood cells to remove red blood cells. The blood sample is mixed with the red blood cell lysate at a ratio of 1:3; the cell suspension after red blood cell lysis is added to the filter membrane ( The filter membrane has a pore size of 5-8um), and undergoes suction filtration treatment. After the suction filtration, carefully remove the filter membrane with tweezers, and place ...
Embodiment 3
[0142] Embodiment 3 The influence of different target gene selections on detection results
[0143] 1. Detection of target gene selection
[0144] In order to test the influence of different target gene selections on the sample detection results, this embodiment designs three experimental groups, wherein experimental group 1 uses all probes of the first and second groups to detect the ROS1 gene; experimental group 3 uses the third , All probes in the fourth group were used to detect the C-met gene; Experimental group 3 used all the probes in the first, second, third and fourth groups to detect ROS1 and C-met gene abnormalities in parallel. The specific test arrangement is shown in Table 6. Synthesis of probes, construction of probe library, probe labeling and experimental steps are as shown in Example 1 and Example 2.
[0145] Table 6 Selection of target genes
[0146]
[0147] 2. Sample testing
[0148] Using the kit prepared by the above design, according to the steps...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com