Method for application of H3K27me3 and demethylase KDM6A/B thereof to nuclear transfer reconstructed embryo of mouse
A technology of demethylase and an application method is applied in the application field of mouse nuclear transfer and reconstructed embryos to achieve the effects of improving blastocyst rate, oocyte reprogramming ability, and blastocyst development rate.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0088] The technical solution of the present patent will be described in further detail below in conjunction with specific embodiments.
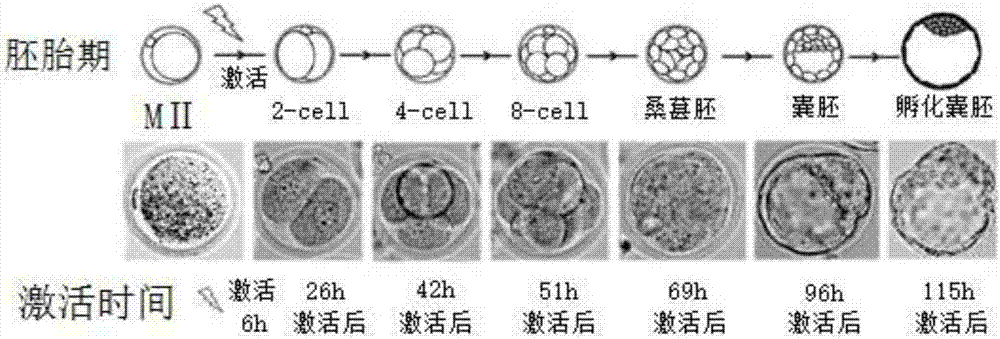
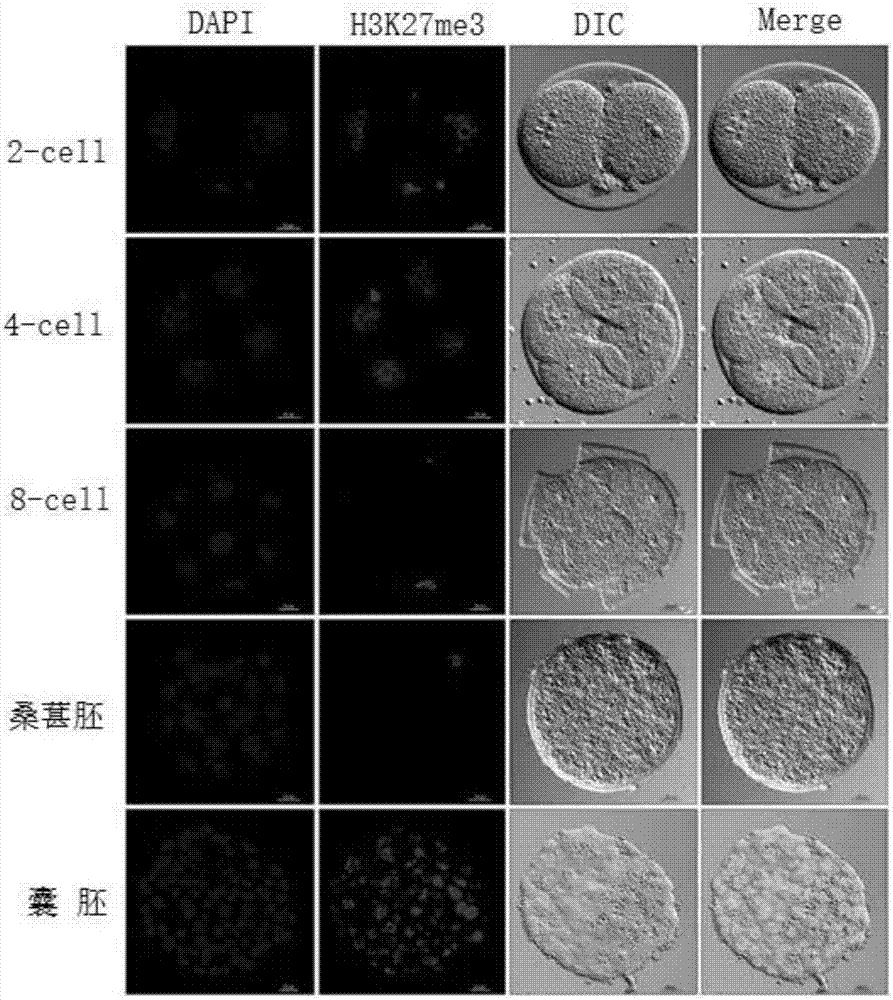
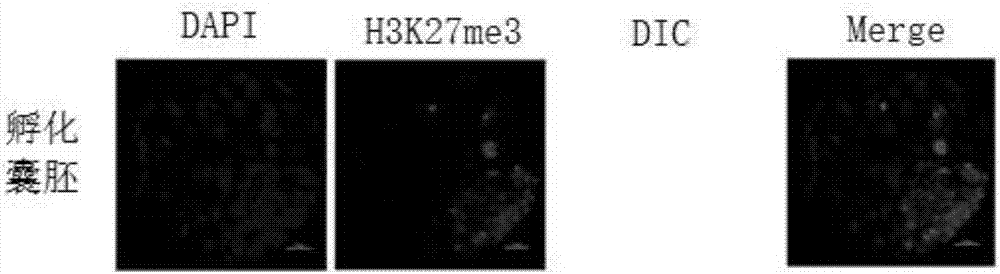
[0089] see Figure 1-30 , an application method of H3K27me3 and its demethylase KDM6A / B in mouse nuclear transfer reconstituted embryos, comprising the following steps:
[0090] 1) Prepare experimental animals:
[0091] Use Oct4 promoter-green fluorescent transgenic mice as oocyte donor mice (OG2) for live cell imaging;
[0092] 2) Collection of MII stage oocytes:
[0093] 3) Prepare experimental reagents:
[0094] 3.1) Embryo culture medium:
[0095] 3.2) Main reagents related to immunofluorescence and western blotting:
[0096] 4% paraformaldehyde (PFA), polyethylene glycol octyl phenyl ether, phosphate buffered saline (PBS), Tween 20, DAPI dye, antibody (Oct4) (dilution ratio 1:500), antibody ( Nanog) (dilution 1:500), Antibody (SSEA1) (1:50 dilution), Antibody (Sox2) (1:500 dilution), Antibody (H3K27me3) (1:50 dilution), Antibody (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com