Primer, fluorescent probe and method for detecting target nucleic acid sequences
A target nucleic acid sequence and fluorescent probe technology, applied in the field of genetic engineering, can solve the problems of interference superposition, affecting the interpretation of melting curve, high fluorescence background, etc., and achieve the effect of clear interpretation and background background reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Embodiment 1 Method for detecting target nucleic acid sequence based on fluorescence energy resonance transfer (FRET)
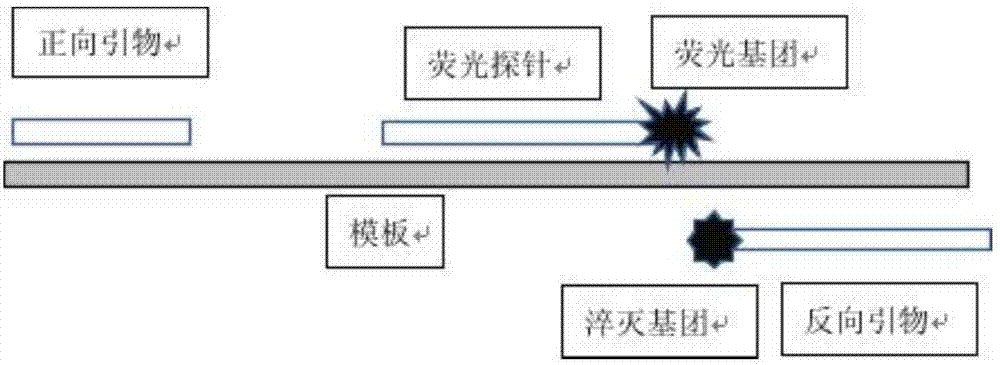
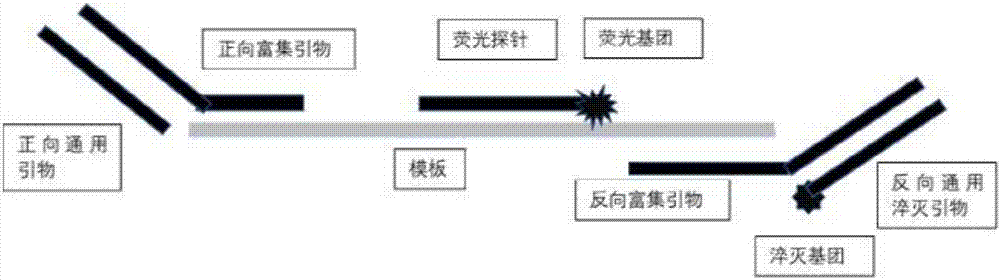
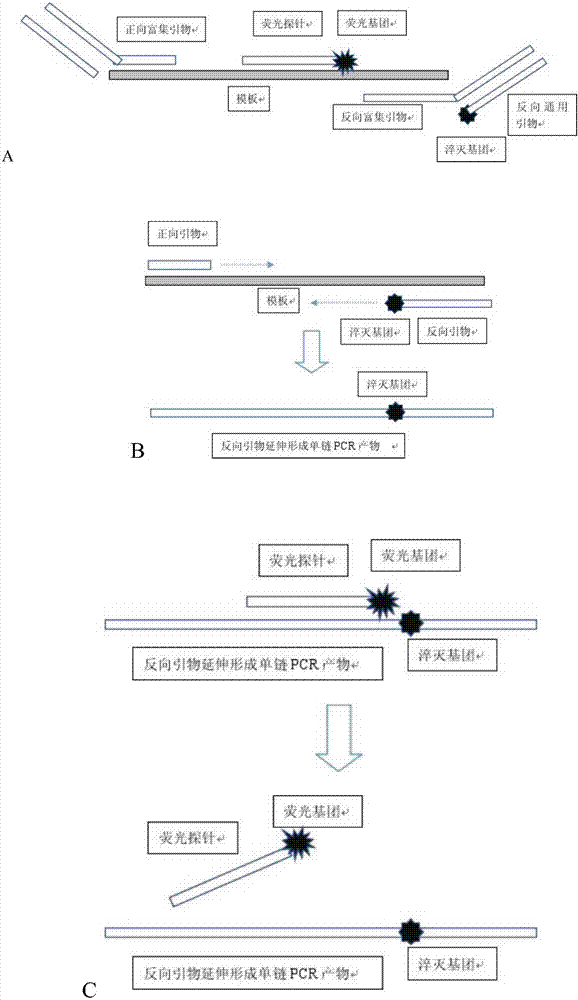
[0029] see Figure 1~2 , is a structural schematic diagram of a primer or a universal primer and a fluorescent probe for detecting a target nucleic acid sequence of the present invention.
[0030] The primer can be designed as a forward primer or a reverse primer. In this example, it is a reverse primer with a quencher group BHQ1 modified near the 3' end. The fluorescent probe is complementary to the different single strands of the primer and DNA, and the near 3' end is modified with the fluorescent group FAM, which can be quenched by the quenching group on the primer, so that the fluorescence intensity decreases ( figure 1 ).
[0031] The 3' end of the forward and reverse enrichment primers is reverse complementary to the template, and the 5' end is the binding site of the universal primer. The universal primer can be designed as a forward primer o...
Embodiment 2
[0039] Example 2 Using Quencher Modified Primers and Fluorescent Group Modified Probes to Detect Different Concentrations of Target Sequence Templates
[0040] According to the principle of Example 1 of the present invention, the method of this example is used to detect different concentrations of target sequence templates to evaluate its practicability.
[0041] The target sequence template is an artificial plasmid containing the PCR amplification region, which was synthesized by Sangon Technology (Shanghai) Co., Ltd. and diluted to 10 3 / ul, 10 4 / ul different concentrations.
[0042] The instrument used is Shanghai Hongshi Slan-96p fluorescence quantitative PCR instrument.
[0043] The primer sequences are:
[0044] STD-F: TCAGCAACTAGCTTACCATC (SEQ ID No: 1), the penultimate T at the 3' end modifies the BHQ1 quenching group
[0045] STD-R: GCTGACGTTGCAAGAAGACG (SEQ ID No: 2)
[0046] The fluorescent probe sequence is:
[0047] STD-P: TGATCATGTAATGGAAGGGGCAAGA (SEQ ID No...
Embodiment 3
[0057] Example 3 Detection of Target Sequence Templates at Different Concentrations Using Fluorophore-Modified Primers and Quencher-Modified Probes
[0058] According to the principle of Example 1 of the present invention, the method of this example is used to detect different concentrations of target sequence templates to evaluate its practicability.
[0059] The target sequence template is an artificial plasmid containing the PCR amplification region, which was synthesized by Sangon Technology (Shanghai) Co., Ltd. and diluted to 10 3 / ul, 10 4 / ul different concentrations.
[0060] The instrument used is Shanghai Hongshi Slan-96p fluorescence quantitative PCR instrument.
[0061] The primer sequences are:
[0062] STD-F: TCAGCAACTAGCTTACCATC (SEQ ID No: 1), the penultimate T at the 3' end
[0063] Modified FAM Fluorophore
[0064] STD-R: GCTGACGTTGCAAGAAGACG (SEQ ID No: 2)
[0065] The fluorescent probe sequence is:
[0066] STD-P: TGTAATGGAAGGGGCAAGA (SEQ ID No: 3), ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com