A gene chip kit for detecting drug-resistant genes of Gram-negative bacteria
A Gram-negative, drug-resistant gene technology, applied in the field of nucleic acid amplification, can solve the problems of cumbersome operation, low throughput, and slow report results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Embodiment 1, preparation and use thereof for detecting the kit of Gram-negative bacterial drug resistance gene
[0062] 1. Assembly and preparation of kits for detecting drug resistance genes of Gram-negative bacteria
[0063] 1. Primers and hybridization probes designed for 10 Gram-negative bacterial drug resistance genes
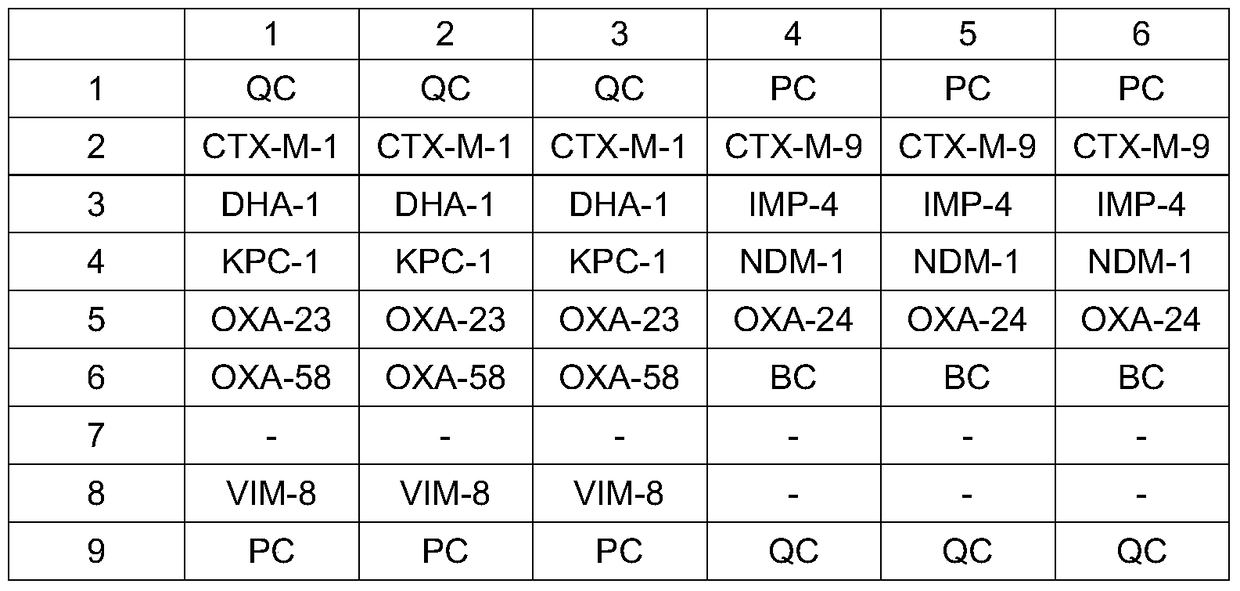
[0064] The specific sequences of primers and single-stranded hybridization probes designed for 10 Gram-negative bacterial drug resistance genes are shown in Table 1 and Table 2.
[0065] Table 1 Primers designed for 10 Gram-negative bacterial drug-resistant genes
[0066]
[0067]
[0068] Table 2 Single-stranded hybridization probes designed for 10 Gram-negative bacterial drug-resistant genes
[0069] Numbering
detection gene
probe name
Sequence (5'-3')
1
bla DHA-1
EF406115.1
DHA-574
TTGCTGACTGCACGGATCCT (sequence 21)
2
bla OXA-23
JN665073.1
OXA23-139
CCCGAGTC...
Embodiment 2
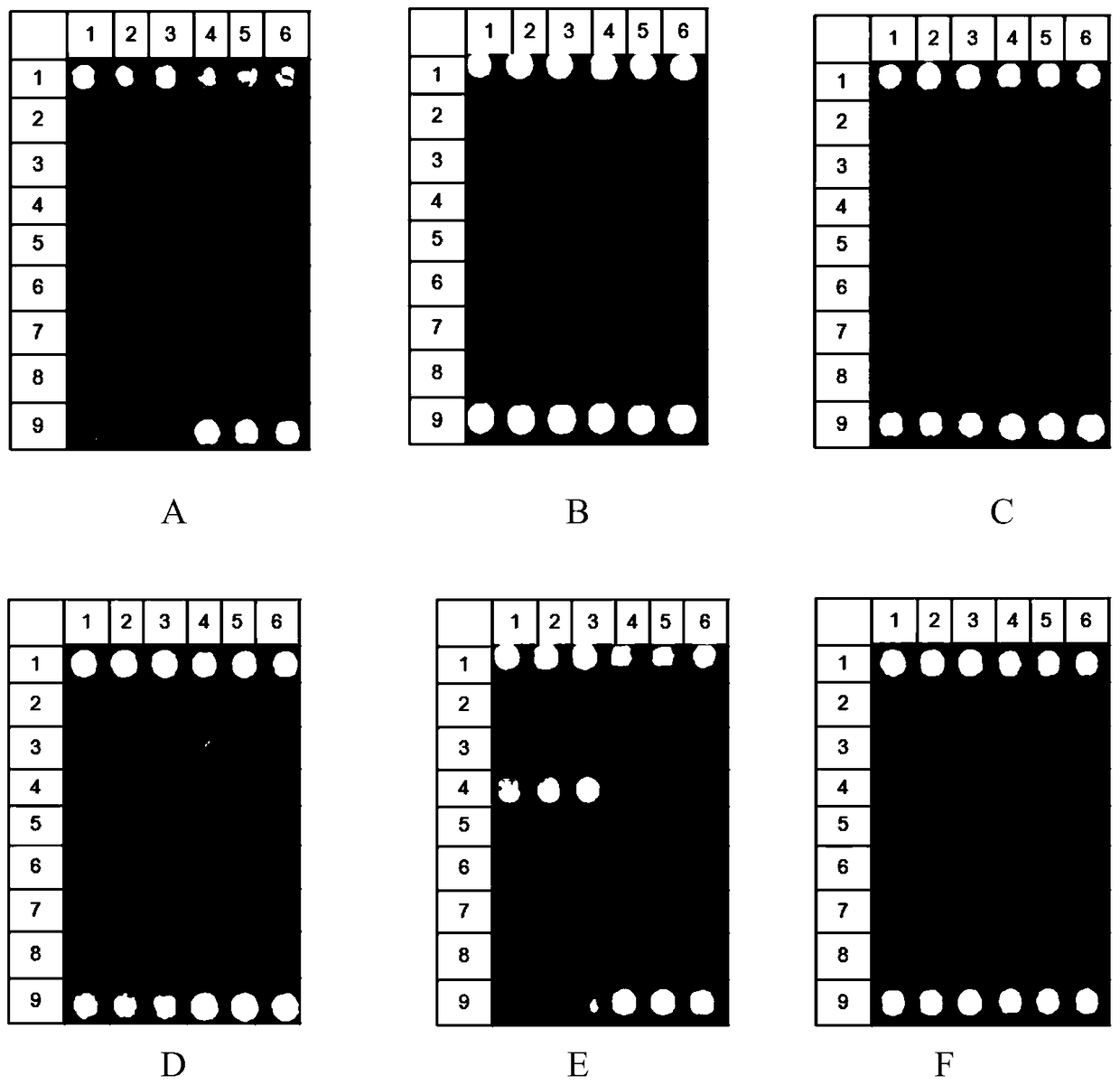
[0091] Embodiment 2, the specificity and the sensitivity determination of the kit for detecting drug resistance gene of Gram-negative bacteria
[0092] 1. Preparation of reference DNA plasmid
[0093] 1. Contains bla DHA-1 Construction of plasmids for gene target fragments
[0094] will bla DHA-1 The DNA fragment shown at positions 1-1113 of the gene (GenBank: EF406115.1, update: 2007-8-17) was ligated to the pGEM-T Easy Vector vector (promega company product) to obtain the recombinant plasmid ZL-DHA-1. and verified by sequencing.
[0095] 2. Contains bla OXA-23 Construction of plasmids for gene target fragments
[0096] will bla OXA-23 The DNA fragment shown at positions 97-899 of the gene (GenBank: JN665073.1, update: 2011-11-7) was ligated to the pGEM-T Easy Vector vector to obtain the recombinant plasmid ZL-OXA-23. and verified by sequencing.
[0097] 3. Contains bla OXA-24 Construction of plasmids for gene target fragments
[0098] will bla OXA-24 The DNA fragm...
Embodiment 3
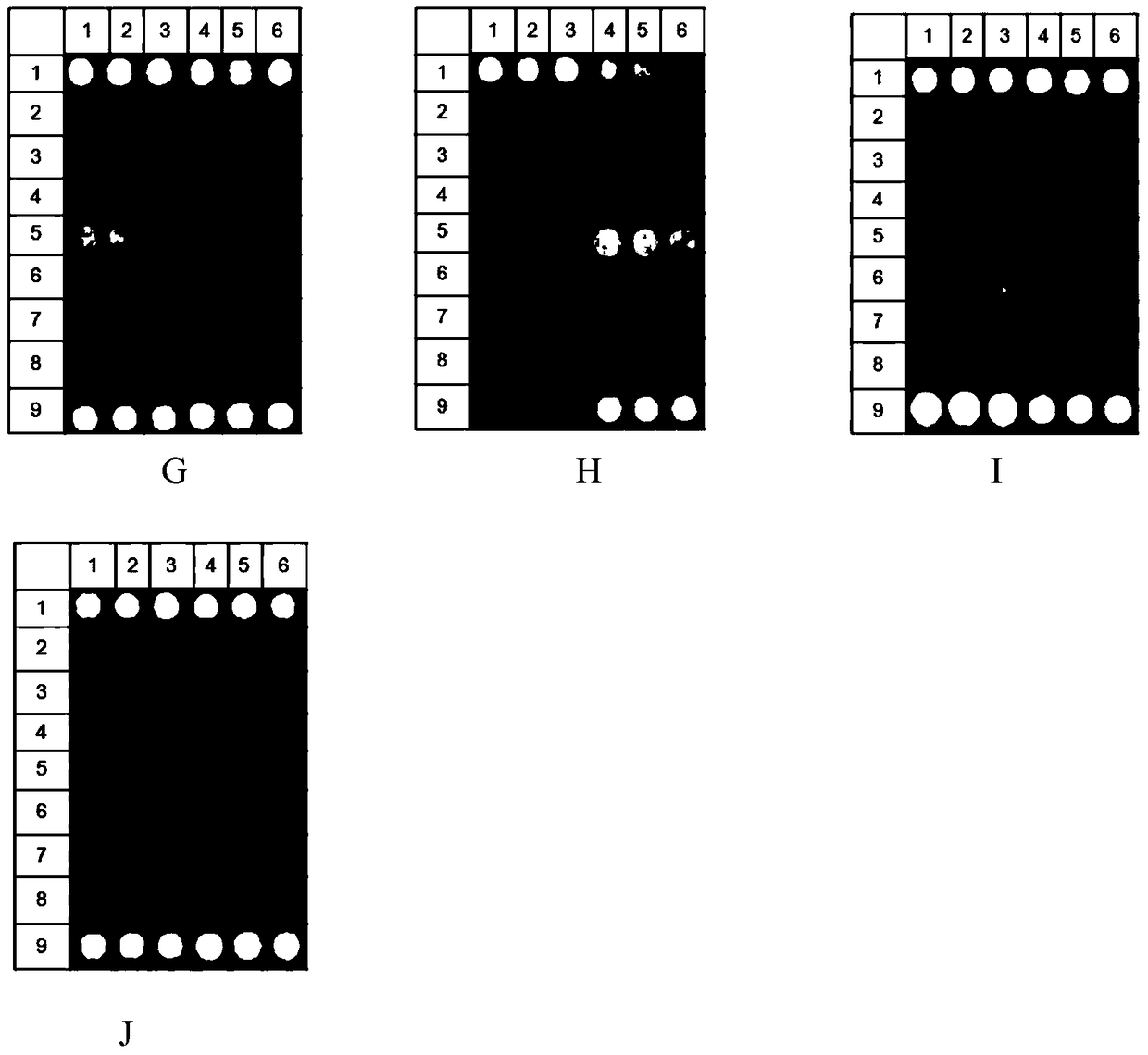
[0119] Embodiment 3, the detection of actual clinical sample
[0120] 1. Types of clinical samples
[0121] The clinical samples used in this example came from viscous pus from human wounds collected by Peking University People's Hospital (based on the principle of voluntariness of the collectors), a total of three samples.
[0122] 2. Extraction of DNA samples from clinical samples
[0123] 1. Take 1-3mL of clinical samples from step 1;
[0124] 2. Add 4 times the volume of 4% (4g / 100mL) NaOH, shake well, and place at room temperature for 30 minutes to liquefy;
[0125] 3. Take 0.5mL of liquefied pus and 0.5ml of 4% (4g / 100mL) NaOH, room temperature, 10min;
[0126] 4. Centrifuge at 12 000 rpm for 15 minutes;
[0127] 5. Discard the supernatant, add 1 mL of sterile saline, mix well, and centrifuge at 12 000 rpm for 5 min;
[0128] 6. Discard the supernatant, and precipitate for DNA extraction;
[0129] 7. Add 50 μL of nucleic acid extraction solution (product of Boao Bi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com