Method for rapidly detecting pseudomonas aeruginosa in textile
A technology for Pseudomonas aeruginosa and textiles, which is applied in the field of detection of Pseudomonas aeruginosa, and can solve the problems of complicated procedures, inability to detect multiple components at the same time, and long cycle.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
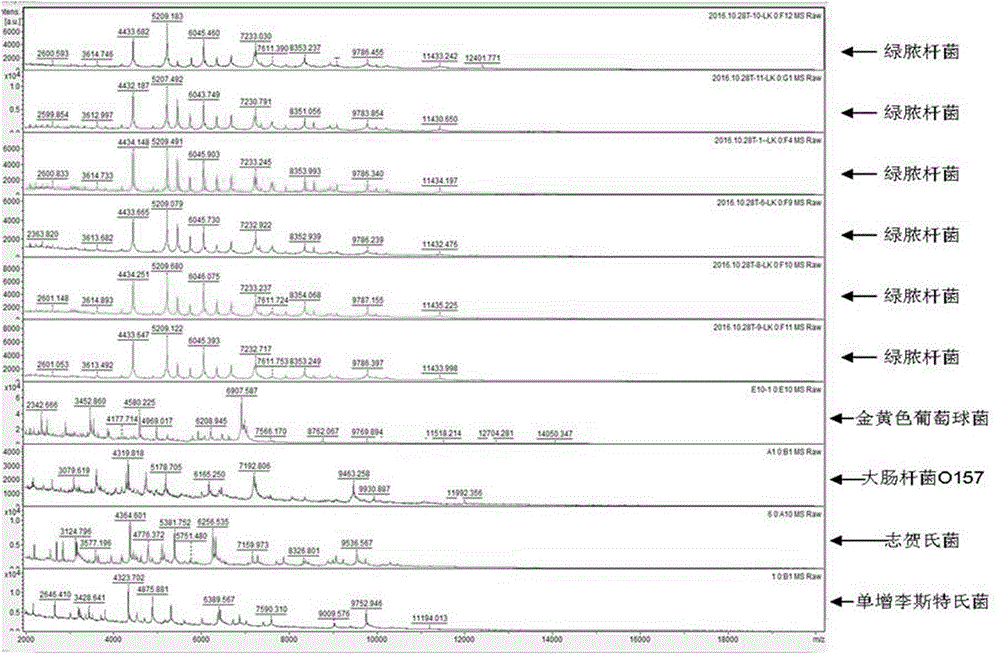
[0099] A method for rapidly detecting Pseudomonas aeruginosa in textiles, comprising the following steps:
[0100] Step 1: Sample enrichment
[0101] Aseptically open the textile sample for inspection, cut the sample evenly with sterile scissors, accurately weigh 25g of the cut sample on an electronic balance, cut it into pieces and add it to a sterile homogeneous bag containing 225mL of SCDLP liquid medium. Beat with a slap-type homogenizer for 1min to 2min, and mix well. Put the prepared samples into a constant temperature incubator at 36°C±1°C, and enrich the bacteria for 24h±2h.
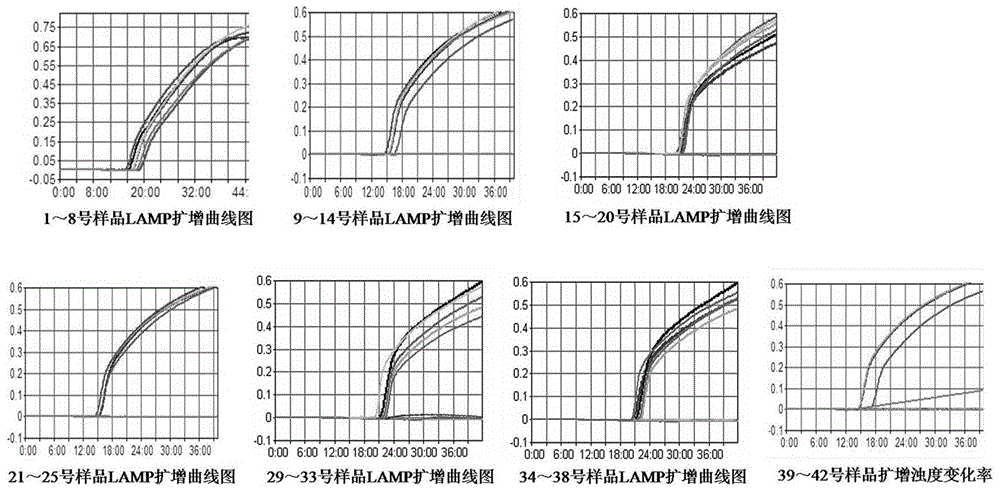
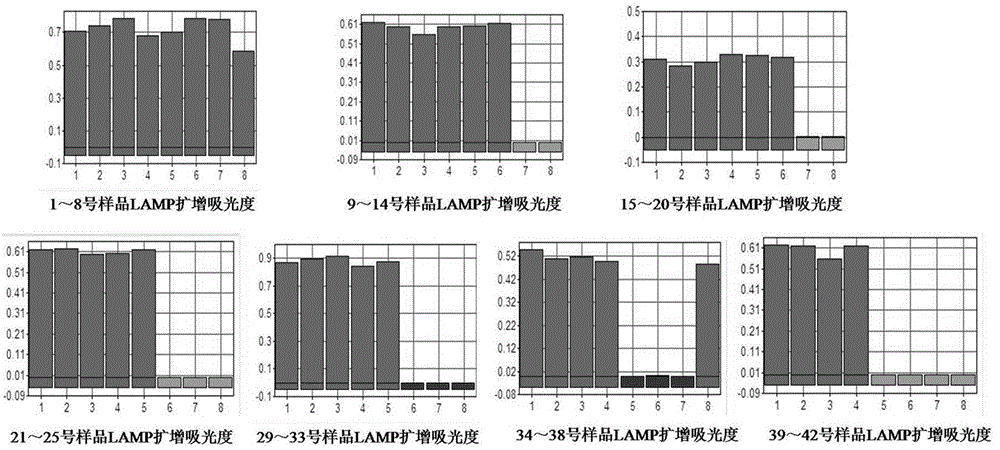
[0102] Step 2: LAMP test
[0103] 2.1) Preparation of DNA template: Take 1mL of SCDLP enrichment solution and put it into a 1.5mL sterile Eppendorf tube, centrifuge at 10000r / min in a high-speed refrigerated centrifuge for 5min, discard the supernatant; add 1mL of sterilized double-distilled water, mix well, and centrifuge in a high-speed refrigerated centrifuge Centrifuge at 10,000r / min in th...
Embodiment 2
[0136] A method for rapidly detecting Pseudomonas aeruginosa in textiles, comprising the following steps:
[0137] Step 1: Sample enrichment
[0138] Aseptically open the textile sample for inspection, cut the sample evenly with sterile scissors, accurately weigh 25g of the cut sample on an electronic balance, cut it into pieces and add it to a sterile homogeneous bag containing 225mL of SCDLP liquid medium. Beat with a slap-type homogenizer for 1min to 2min, and mix well. Put the prepared samples into a constant temperature incubator at 36°C±1°C, and enrich the bacteria for 24h±2h.
[0139] Step 2: LAMP test
[0140] 2.1) Preparation of DNA template: Take 1mL SCDLP enrichment solution and put it into a 1.5mL sterile Eppendorf tube, centrifuge in a high-speed refrigerated centrifuge at 10,000r / min for 5min, discard the supernatant; add 1mL sterilized double-distilled water to mix, and freeze at high speed Centrifuge at 10,000r / min in a centrifuge for 5min, discard the super...
Embodiment 3
[0172] A method for rapidly detecting Pseudomonas aeruginosa in textiles, comprising the following steps:
[0173] Step 1: Sample enrichment
[0174] Aseptically open the textile sample for inspection, cut the sample evenly with sterile scissors, accurately weigh 25g of the cut sample on an electronic balance, cut it into pieces and add it to a sterile homogeneous bag containing 225mL of SCDLP liquid medium. Beat with a slap-type homogenizer for 1min to 2min, and mix well. Put the prepared samples into a constant temperature incubator at 36°C±1°C, and enrich the bacteria for 24h±2h.
[0175] Step 2: LAMP test
[0176] 2.1) Preparation of DNA template: Take 1mL of SCDLP enrichment solution and put it into a 1.5mL sterile Eppendorf tube, centrifuge at 10000r / min in a high-speed refrigerated centrifuge for 5min, discard the supernatant; add 1mL of sterilized double-distilled water, mix well, and centrifuge in a high-speed refrigerated centrifuge Centrifuge at 10,000r / min in th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com