A pig SNP marker site analyzing method based on a sequence-based typing technique
An analysis method and genotyping technology, applied in the field of analysis of pig SNP marker sites, can solve problems such as difficult breeds or strains, inapplicability, etc., achieve the effects of reducing typing costs, improving sequencing data quality, and broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Prediction of EcoRI-MspI Double Enzyme Cutting Porcine Genome Effect
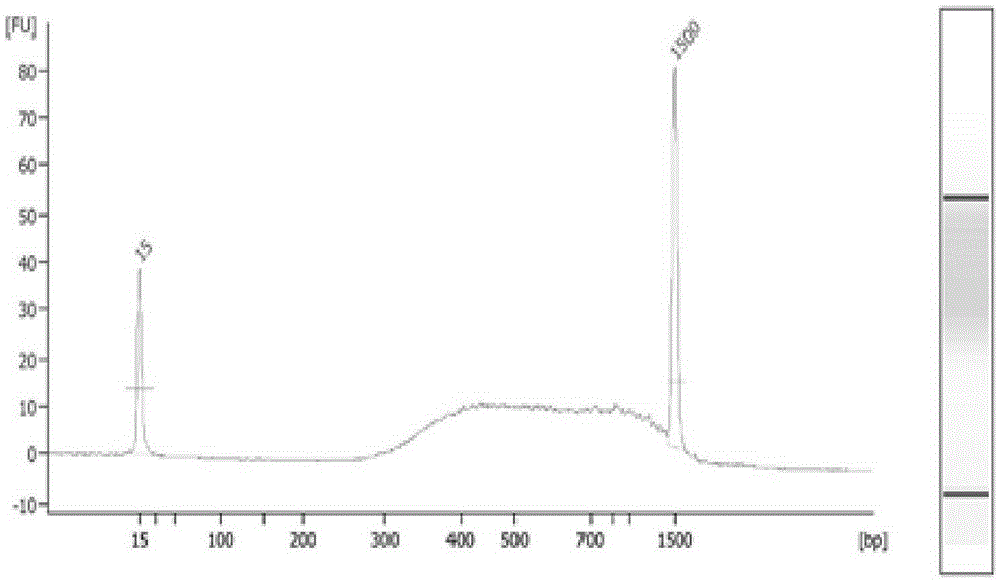
[0037] This example is used to illustrate the number and size distribution of restriction fragments obtained by EcoRI-MspI double digestion combination. Bioinformatics analysis method was used to simulate the digested pig genome, and the number of double-digested fragments in the range of 100-300bp was counted. The results showed that about 135,495 fragments with a size of 100-300bp could be obtained when EcoRI-MspI double-digested the pig genome Double restriction fragments, which are suitable for simplified genome sequencing library construction are utilized.
Embodiment 2
[0042] Example 2 Simplified Genome Sequencing of Multiple Pig Breeds Using EcoRI-MspI Digestion
[0043] This example is used to illustrate that pig genome can be digested with EcoRI-MspI to obtain SNP information covering the whole genome.
[0044] 1. Experimental materials
[0045] Ear tissue samples of Landrace pigs, Large White pigs, Erhualian pigs, Shawutou pigs, Meishan pigs, and Mi pigs were collected, with 4 pigs for each breed, totaling 24 pig ear tissue samples. Genomic DNA was extracted with DNeasy Blood&Tissue Kit, and the genome concentration was diluted to 50ng / μL for later use.
[0046] 2. Design universal adapters (including oligo1 and oligo2), barcode adapters (including oligo1 and oligo2), and PCR amplification primers (including oligo1 and oligo2) according to the restriction recognition sequences of EcoRI and MspI. There are 24 barcode adapters in total for The 24 samples were differentiated so that they could be sequenced together. Sequences were synthe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com