Quick and efficient detection method of mutation sites of genes associated with corneal dystrophy
A technology for malnutrition and mutation sites, applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of slow speed, low sensitivity, and inflexible experiments.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Embodiment 1 mutation DNA template preparation and primer design:
[0044] In order to simulate real cases in the laboratory, we artificially synthesized TGFBI gene fragments containing 4 mutation sites to test the detection efficiency of the mutation sites. At the same time, we designed specific amplification primers for the normal site and the mutant site (the two primers differ only in the terminal bases, respectively denoted by F1 and F2), and the corresponding reverse primers (represented by R), 3 Primer combinations form a primer master mix. In addition, the 5' ends of the two forward primers are respectively connected with different detection primer sequences for fluorescence detection.
[0045] The primer sequences of the specific 4 sites are as follows:
[0046] TGFBI-371G-A
[0047] F1: 5′-GAAGGTGACCAAGTTCATGCTCACTCAGCTGTACACGGACCG-3′
[0048] F2: 5′-GAAGGTCGGAGTCAACGGATTCACTCAGCTGTACACGGACCA-3′
[0049] R: 5′-CCCCTCCATCTCAGGCCTCA-3′
[0050] TGFBI-1526T-C...
Embodiment 2
[0062] The establishment of embodiment 2 PCR detection system:
[0063] The PCR amplification system includes commercially purchased 2x Mix (including Taq enzyme, 4 kinds of dNTPs, and two probes corresponding to the detection primer sequence of the forward primer). The PCR system is as follows:
[0064] PCR amplification system (10μL):
[0065]
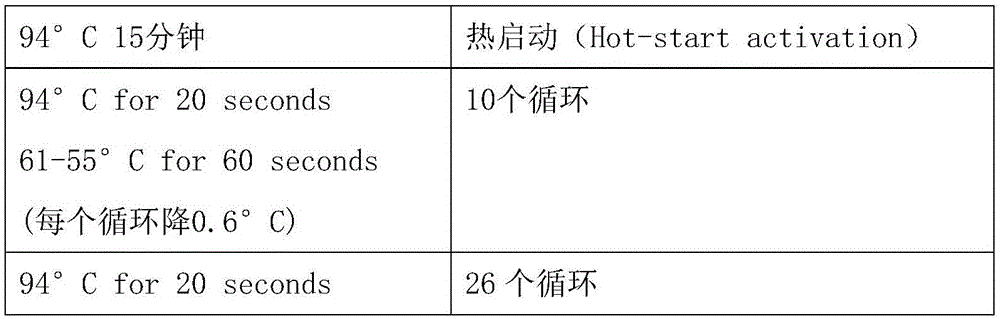
[0066] The PCR amplification procedure is as follows:
[0067]
[0068] Fluorescence reading:
[0069] In an environment below 40°C, read the fluorescence value.
[0070] Interpretation of results:
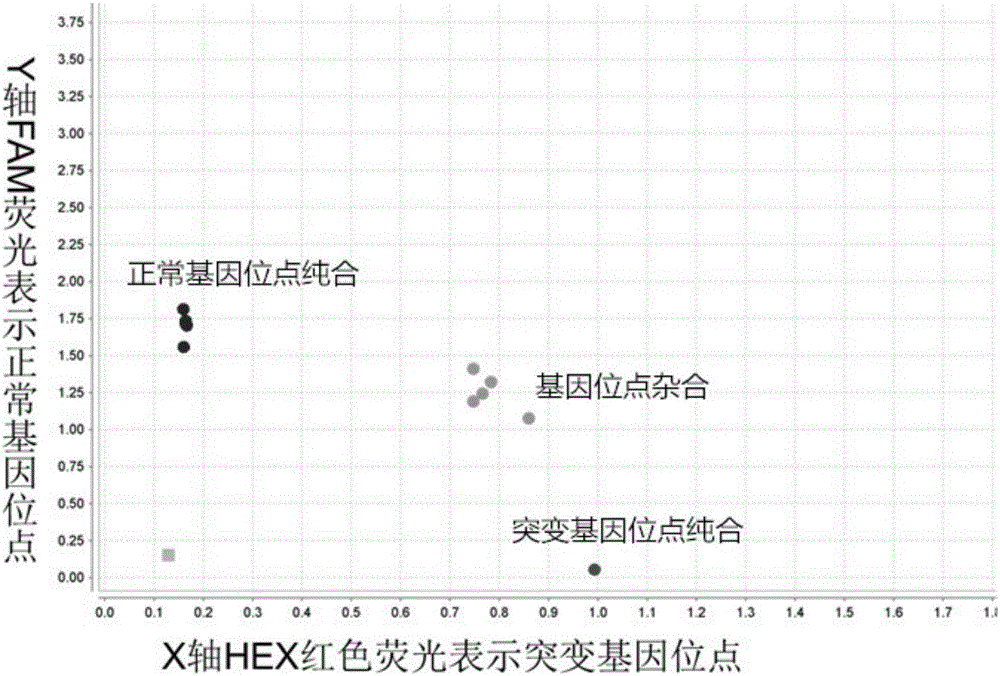
[0071] If the individual contains only normal gene loci, only the specific primers of the normal loci can amplify, and the corresponding fluorescent signal is blue, that is, only blue fluorescence can be read when the fluorescence value is read, indicating that the individual is Homozygous for the normal site; if the individual only contains the mutated gene site, only the specific primers for the mutated site can be amplified...
Embodiment 3
[0072] Example 3 Clinical sample collection and verification:
[0073] A total of 80 clinical samples were collected, and the accuracy of the detection method was analyzed. First, through the above method, 80 clinical samples were detected for mutation sites, and the detection results are shown in Table 1. Further sequencing verification was performed on the discovered mutation sites, and the results showed that the sequencing results were completely consistent with the quantitative PCR detection results. It shows that the method can achieve 100% accuracy in the detected 80 clinical samples, and can be used for clinical gene detection.
[0074] Table 1. Test results for 80 samples.
[0075]
[0076] It can be seen that compared with the existing technology, this technology has the advantages of accurate and reliable detection results, high throughput, flexible experimental design, simple and fast operation, fast detection speed, easy promotion, and low cost.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com