Series G-quadruplex-heme DNA enzyme label-free signal amplification method based on rolling-circle amplification
A technology of rolling circle amplification and signal amplification, applied in the field of molecular biology, which can solve the problems of requiring special markers, relying on large instruments, and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0087] Feasibility Analysis:
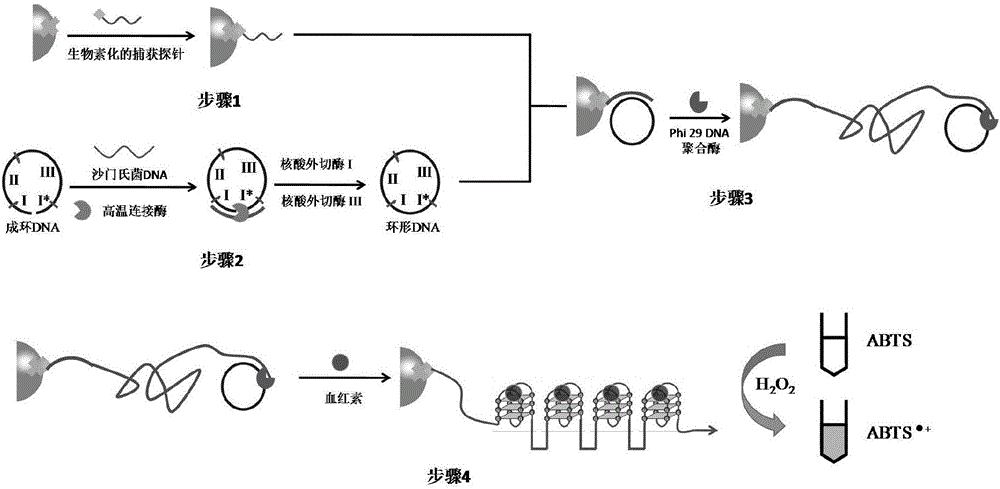
[0088] Preparation of pretreatment microspheres: take 100 μL streptavidin functionalized microspheres, wash twice with 100 μL affinity eluent, centrifuge at 3500 rpm for 5 min, remove the supernatant, add 49.4 μL affinity eluent and 0.6 μL 100 μmol / L of biotinylated capture probe was shaken at 37°C for 1 hour, centrifuged and washed twice with affinity eluent to remove unbound probes, and finally 100 μL of affinity eluent was added and incubated at 4°C Save the pretreated beads.
[0089] Preparation of circularized DNA: Take a sterilized dry centrifuge tube, measure 74.5 μL of deionized water with a pipette gun, add 10 μL of 10× ligase reaction buffer solution, 5 μL of 5’ phosphorylated 1 μmol / L of circular DNA, 10 μL of synthetic Salmonella DNA sequence (10 pmol / L), 0.5 μL of high temperature ligase in a total volume of 100 μL. After the solution was mixed, the ligation cycle amplification technique was adopted, and the steps were as follows:...
Embodiment 2
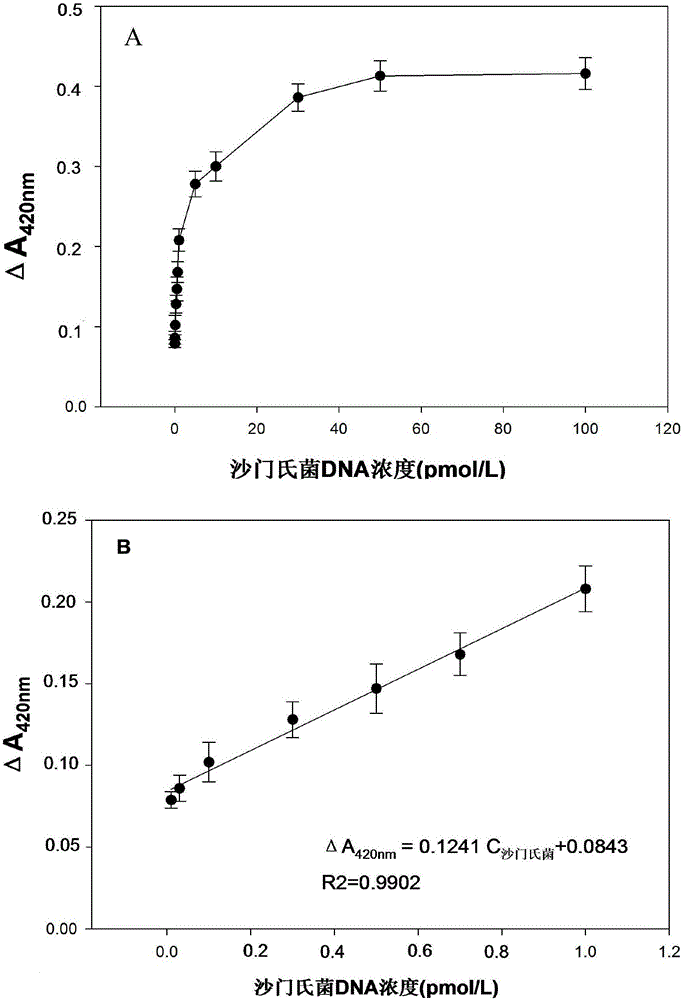
[0093] sensitivity analysis:
[0094] Preparation of pretreated microspheres: take 100 μL of streptavidin functionalized microspheres and wash twice with 100 μL of affinity eluent, centrifuge at 3500 rpm for 5 min, remove the supernatant, add 49.4 μL of affinity eluent and 0.6 μL of 100 μmol / L biotinylated capture probe was shaken at 37°C for 1 hour, centrifuged and washed twice with affinity eluent to remove unbound probes, and finally 100 μL of affinity eluent was added and incubated at 4°C Save the pretreated beads.
[0095]Preparation of circularized DNA: Take a sterilized dry centrifuge tube, measure 74.5 μL of deionized water with a pipette gun, add 10 μL of 10× ligase reaction buffer solution, 5 μL of 5’ phosphorylated 1 μmol / L of circular DNA, 10 µL of synthetic Salmonella DNA-seq, 0.5 µL of high-temperature ligase in a total volume of 100 µL. After the solution was mixed, the ligation cycle amplification technique was adopted, and the steps were as follows: place t...
Embodiment 3
[0098] Performance analysis for the detection of Salmonella in real samples:
[0099] Preparation of pretreated microspheres: 100 μL of streptavidin functionalized microspheres were washed twice with 100 μL of affinity eluent, and centrifuged at 3500 rpm for 5 min. Remove the supernatant, add 49.4 μL of affinity eluent and 0.6 μL of 100 μmol / L biotinylated capture probe, shake at 37°C for 1 hour, centrifuge and wash twice with affinity eluent to remove unbound Finally, add 100 μL of affinity eluent and store the pretreated microspheres at 4°C.
[0100] Cultivation of Salmonella and digestion of genomic DNA: Salmonella was cultured in LB medium, and the genomic DNA of Salmonella was extracted using a commercial general bacterial genomic DNA extraction kit. Treat the extracted Salmonella genomic DNA with RsaI restriction endonuclease, the conditions are: add 10U of RsaI and its reaction buffer solution to 50ng of Salmonella genomic DNA, with a total volume of 50μL, react at 37°...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com