LNA probe for detecting KRAS gene mutation and detection method
A detection method and probe technology, applied in the field of KRAS gene mutation detection, can solve the problems of time-consuming, high background of DNA sequencing, false positives or false negatives, etc., and achieve simple operation, high single-base resolution, Specificity-enhancing effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Embodiment 1: the establishment of detection method
[0038] 1. Design corresponding LNA probes for the three mutant sequences of the 12 and 13 codon mutations of the KRAS gene.
[0039] The KRAS mutant gene sequence is as follows:
[0040] name Sequence (5'-3') Types of 135A GTT GGA GCT GAT GGC GTA G dna 135C GTT GGA GCT GCT GGC GTA G dna 134T GTT GGA GCT TGT GGC GTA G dna
[0041] The probe sequences are as follows:
[0042] name Sequence (5'-3') Types of position in the sequence listing LNA-P 135A
ACG CCA TCA GCT C LNA sequence 1 LNA-P 135C
ACG CCA GCA GCT C LNA sequence 2 LNA-P 134T
ACG CCA CAA GCT C LNA sequence 3
[0043] 2. The LNA probes were hybridized with the corresponding three kinds of KRAS mutant genes, and three kinds of hybrids were obtained.
[0044]The LNA probe and the KRAS mutant gene were respectively made into 100 μM stock solution with ult...
Embodiment 2
[0050] Embodiment 2: Utilize LNA probe to detect KRAS mutant gene
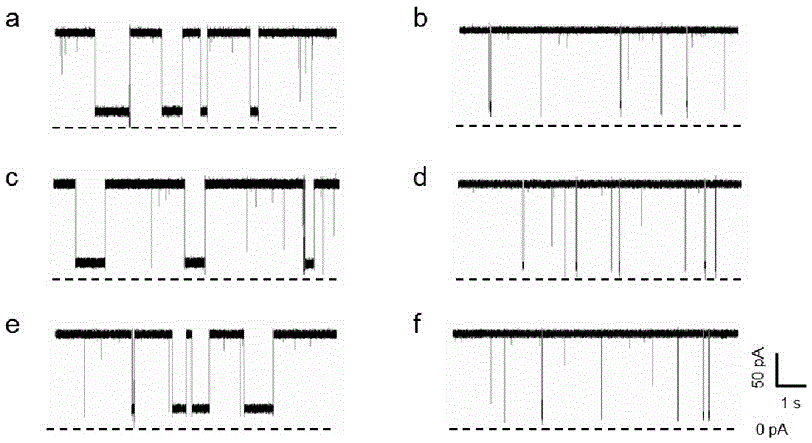
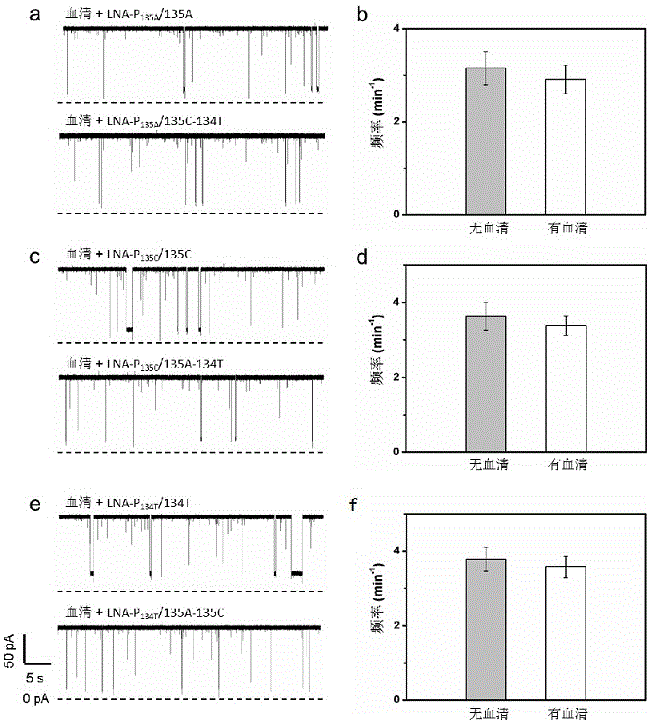
[0051] The three kinds of probes were hybridized with the corresponding KRAS mutant genes respectively, and the three kinds of hybrids obtained (135A / LNA-P 135A , 135C / LNA-P 135C and 134T / LNA-P 134T ) were detected by nanopore sensors, respectively. As a control, the probes were hybridized with the mixture of the other two mutated genes, and detected with nanopore sensors under the same conditions (voltage: +120mV, electrolyte solution: 1M KCl (pH=7.8, 10mM Tris-HCl, 1mM EDTA)) , the result is as figure 1 shown. LNA-P 135A Hybrid with mutant gene 135A (135A / LNA-P 135A ) was injected into the detection pool, a long time blocking event was generated ( figure 1 a), the fitting value of blocking time is 452.17ms, while the probe LNA-P 135A After the mixture with the other two mutant genes was detected by the nanopore, the blocking signal was particularly short, and the blocking time was 23.55ms ( figure 1...
Embodiment 3
[0052] Example 3: Quantitative analysis of the KRAS mutant gene
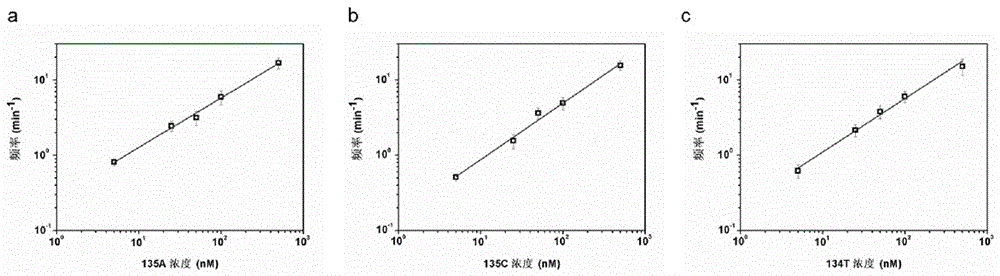
[0053] Taking the KRAS mutant gene 135A as an example, different concentrations of 135A and equal concentrations of the probe LNA-P 135A After hybridization, the final concentrations after injection into the detection pool were 5nM, 25nM, 50nM, 100nM, and 500nM, and the frequency of blocking events was counted, and the relationship between the frequency and the concentration of the mutant gene 135A was established, so as to perform quantitative analysis on 135A. The result is as figure 2 as shown in a. The quantitative analysis results of 135C and 134T are shown in figure 2 b and figure 2 c. The three mutant genes showed a good linear relationship between the detection concentration of 5nM-500nM.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com