Kit for human papilloma virus E6/E7 gene detection and detection method
A human papillomavirus and gene detection technology, applied in the fields of life sciences and methods, can solve the problems of decreased sensitivity of detection of HPV, deletion of HPVL1 gene, and difficulty in grasping the amount of sample DNA.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0131] 1. Sample DNA extraction
[0132] Take 200uL of the cervical exfoliated cell suspension to be tested and put it into a 1.5mL tube, centrifuge at 13000r / min for 5min, discard the supernatant, add 200uL of nucleic acid extraction solution to the precipitate, shake and mix well, lyse at 1000C for 10min, centrifuge at 13000r / min for 5min, take the upper 3uL of the supernatant was used as a template for the PCR reaction.
[0133] 2. Multiplex PCR reaction
[0134] The PCR reaction template is added to the reaction system, the reaction system is 10uL: DDH 2 O 2.35uL, 10*PCR Buffer1uL, 25nM Mg 2+ 1uL, 1.5uL of 2nM DNTP, 1uL of primer mixture (i.e. the primers shown in Table 1, the working concentration of each primer is 0.33uM), 0.15uL of 5U / uL FastTaq enzyme, 3uL of sample DNA (PCR reaction template). PCR cycle program: 95°C 4min; 11 cycles x (94°C 30s, 68°C-0.5°C / cycle 90s); 28cycles x (94°C 30s, 63°C 90s); 72°C 5min; 4°C for ever.
[0135] The multi-cycle reaction prog...
Embodiment 2
[0142] 1. Sample DNA extraction
[0143] Take 200uL of the cervical exfoliated cell suspension to be tested, use the automatic nucleic acid extractor NP-968 nucleic acid automatic extractor to extract DNA according to the instructions, and take 3uL as a PCR reaction template.
[0144] 2. Multiplex PCR reaction
[0145] The PCR reaction template is added to the reaction system, the reaction system is 10uL: DDH 2 O 2.35uL, 10*PCR Buffer1uL, 25nM Mg 2+ 1uL, 1.5uL of 2nM DNTP, 1uL of primer mixture (ie, the primers shown in Table 1, the working concentration of each primer is 0.33uM), 0.15uL of 5U / uL FastTaq enzyme, and 3uL of sample DNA. PCR cycle program: 95°C 4min; 11 cycles x (94°C 30s, 68°C-0.5°C / cycle 90s); 28cycles x (94°C 30s, 63°C 90s); 72°C 5min; 4°C forever.
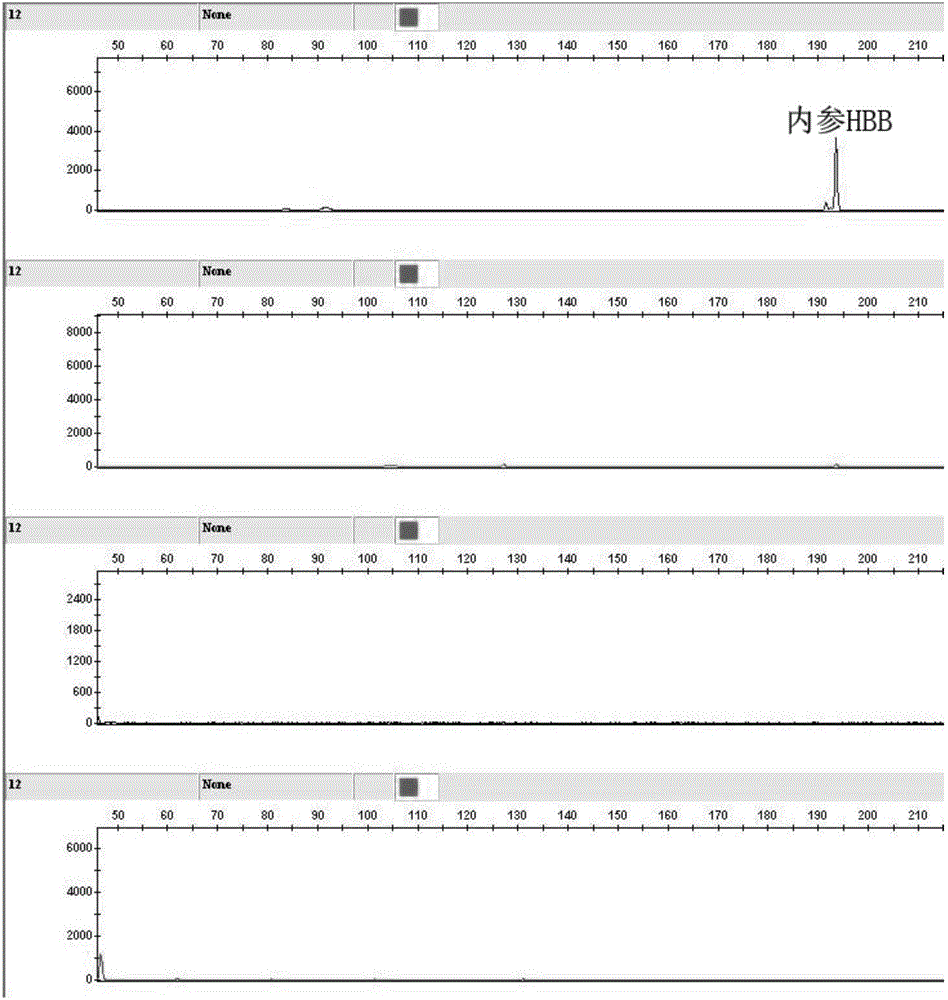
[0146] 3. Genetic analyzer on PCR products
[0147] Take 1uLPCR product using DDH 2 O was diluted 50 times, and 1uL of the diluted solution was mixed with 0.06μl Liz120SIZESTANDARD and 8.9μl Hi-Di, and put on ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com