LAMP method for detecting enterobacteriaceae food-borne pathogenic bacteria, nucleic acid and primer pairs
A technology for the detection of food-borne pathogenic bacteria and Enterobacteriaceae, which is applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., and can solve problems such as low counting results, high counting results, and long detection cycles , to achieve accurate and reliable results, low detection cost and fast detection speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Embodiment 1: Enterobacteriaceae PCR detection method specifically comprises the following steps:
[0022] Step 1, designing primers according to the conserved sequences in the genomic DNA sequences of Enterobacteriaceae;
[0023] Through comparative genome and bioinformatics analysis, a specific DNA sequence was found in the Enterobacteriaceae genome DNA sequence, and it was used as a detection target for Enterobacteriaceae. The base sequence of the nucleic acid is:
[0024] 5’—GCCTTTGTGCATTTTACGGAATTTTGTACGCTTTGGTTGTAACATCAGCGACGCTCCTTATTTACGGCCTTTACGCTGCTGCTTTTTCGGTTGAGCAGCCGGTTTTTCCGGTTGTTCAACAGCAGCCATACCACCCAGGATCTCGCCTTTGAAGATCCACACTTTAACGCCGATTACACCATAAGTGGTGTGCGCTTCAGAGGTGTTGTAGTCGATGTCAGCACGCAGAGTGTGCAGAGGTACGCGACCTTCGCGGTACCATTCGGTACGTGCGATTTCCGCGCCGCCCAGACGGCCGCTAACTTCAACTTTGATACCTTTAGCGCCCAGACGCATGGCGTTCTGTACAGCACGCTTCATCGCACGACGGAACATCACGCGACGTTCCAGCTGAGAAGTGATGCTGTCAGCAACCAGTTTAGCGTCCAGTTCAGGTTTACGAACTTCGGCGATATTGATCTGTGCAGGAACGCCAGCGATATCCGCTACGACCTTACGCA—...
Embodiment 2
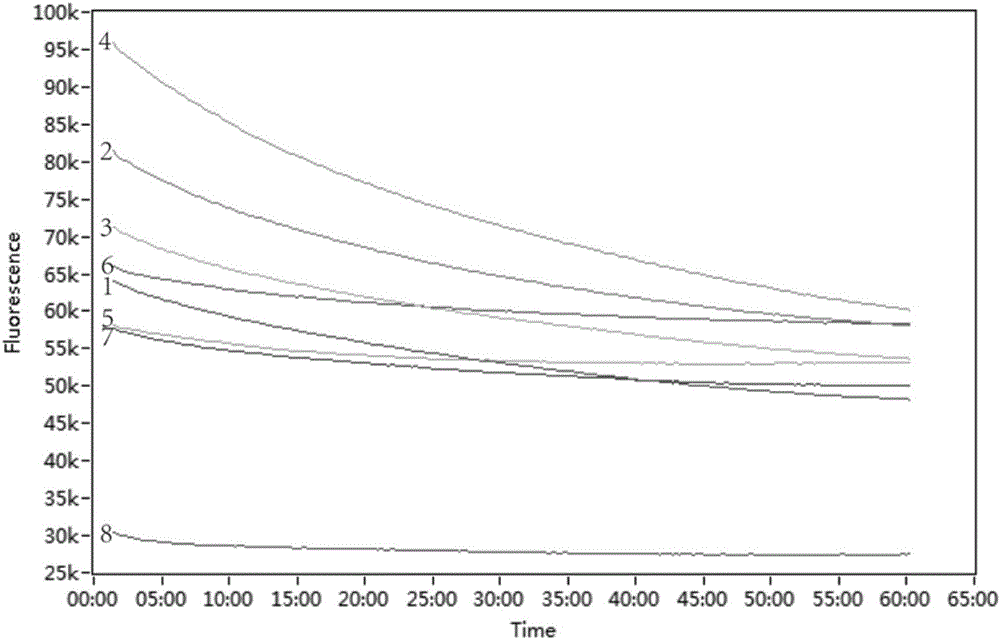
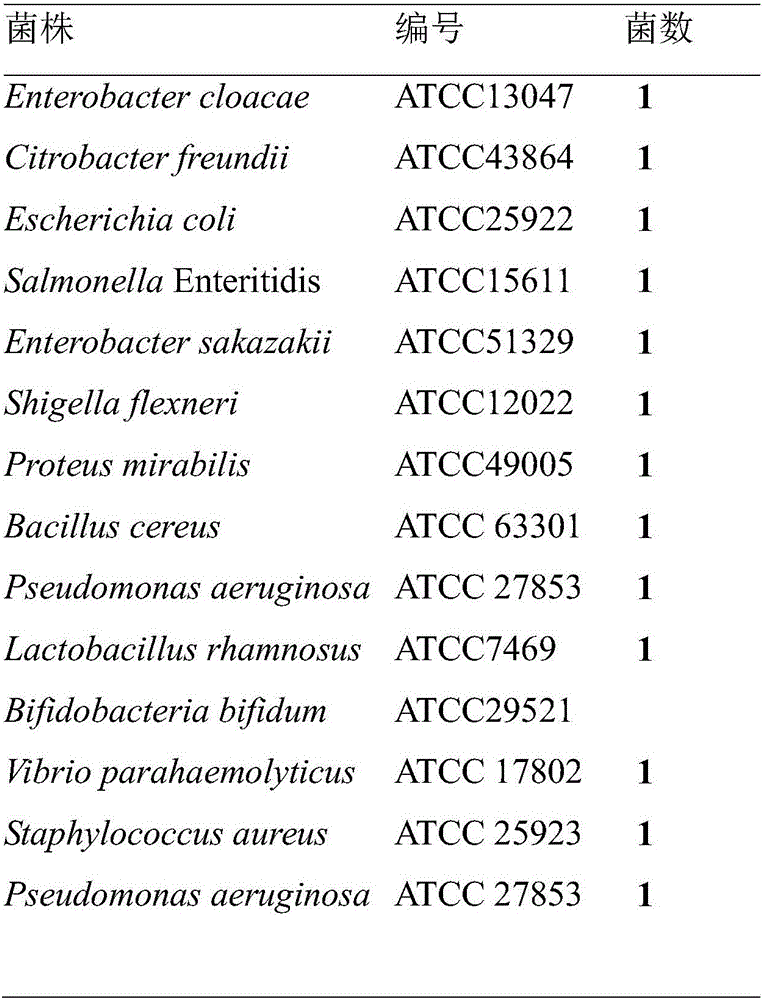
[0044] Example 2: Using the detection method in Example 1, 30 isolated strains from food samples were detected. For sample processing and strain isolation and identification, refer to the national standard GB / T4789.4-2010. The results showed that the LAMP detection was consistent with the identification of the national standard method (as shown in Table 2).
[0045] Table 2 The isolated strains used for detection
[0046]
[0047]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com