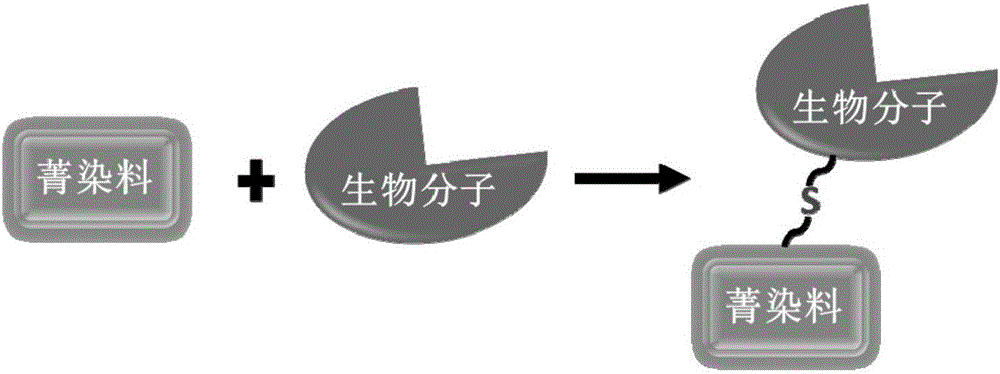
Biomolecule fluorescence marking method, fluorescence-marked biomolecule obtained thereby and application of fluorescence-marked biomolecule
A biomolecular and fluorescent labeling technology, applied in fluorescence/phosphorescence, chemical instruments and methods, analytical materials, etc., can solve the problems of poor water solubility, increase the difficulty of fluorescent dye synthesis and separation, and affect the activity of biomolecules, and achieve efficient labeling. , Avoid the influence of biomolecular activity, the effect of the method green environmental protection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] The structural formula of the cyanine dye compound used in this embodiment is
[0040]
[0041] where R 1 for -(CH 2 ) 5 COOH, R 2 is Cl, n=1.
[0042] Dissolve 1.0 μmol of glutathione (GSH) in a buffer solution of Tris-HCl (pH=7.4), and then add it into a buffer solution of 1.0 μmol of cyanine dye Tris-HCl (pH=7.4), avoid light , stirring at room temperature for 2 hours, after the reaction was completed, the reaction solution was added into a dialysis bag for dialysis separation, and then the solution in the dialysis bag was taken out and freeze-dried to obtain fluorescently labeled GSH. The molecular weight was identified by MALDI-TOF.
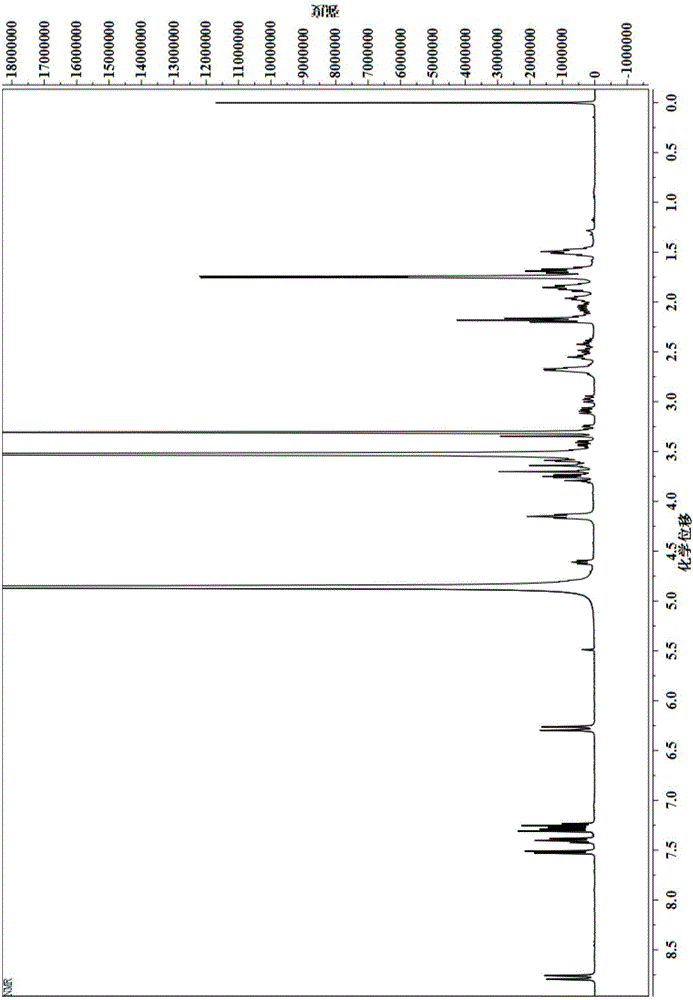
[0043] By H NMR spectroscopy (such as figure 2 Shown) the GSH that gained fluorescent label is good has been characterized, and the result is as follows:
[0044] 1 HNMR (400MHz, CD 3OD, 298K, TMS): δ = 8.79 (d, J = 12Hz, 2H, CH); δ = 7.53 (d, J = 8Hz, 2H, ArH); δ = 7.40 (t, J = 8Hz, 2H, ArH ); 7.31-7.23 (m, 4H, ArH); 6....
Embodiment 2
[0051] The structural formula of the cyanine dye compound used in this embodiment is where R 1 for -(CH 2 ) 5 COOH, R 2 is Cl, n=1.
[0052] Dissolve 1.0 μmol of the polypeptide ECGD (sequence Glu-Cys-Gly-Asp) in Tris-HCl (pH=7.4) buffer solution, and then add 2.0 μmol of cyanine dye Tris-HCl (pH=7.4) In the buffer solution, avoid light, and stir at 20°C for 6 hours. After the reaction is completed, the reaction solution is added to the dialysis bag for dialysis separation, and then the solution in the dialysis bag is taken out and freeze-dried to obtain the fluorescently labeled polypeptide ECGD. The molecular weight was identified by MALDI-TOF.
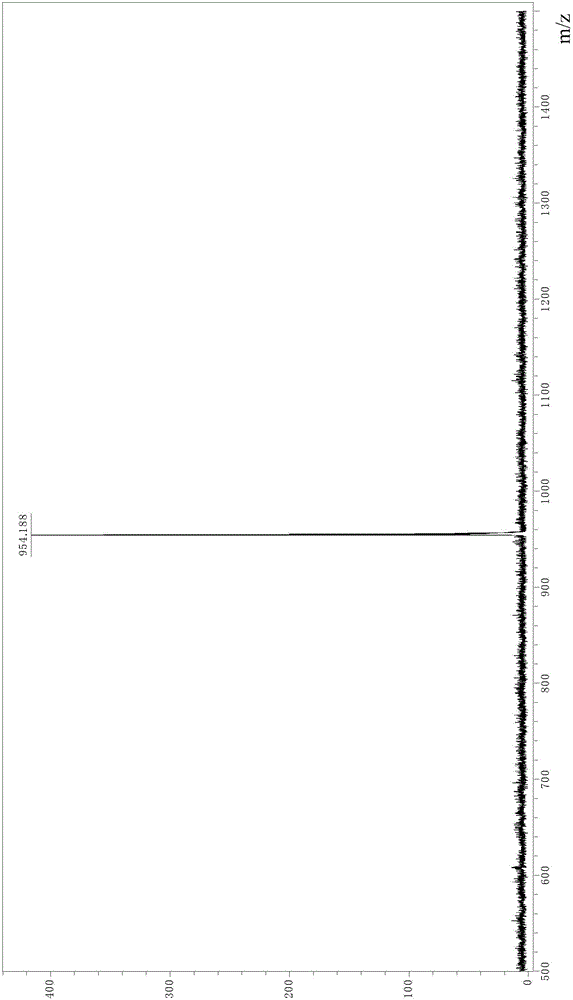
[0053] MALDI-TOF: Calculate C 56 h 73 N 6 o 13 S + The m / z is 1069.49, from Figure 6 The m / z obtained in the spectrum shown was 1069.6.
Embodiment 3
[0055] The structural formula of the cyanine dye compound used in this embodiment is where R 1 for -(CH 2 ) 5 COOH, R 2 is Cl, n=1.
[0056] Dissolve 1.0 μmol of the polypeptide DGRKLVFFCG (sequence Asp-Gly-Arg-Lys-Leu-Val-Phe-Phe-Cys-Gly) in a Tris-HCl (pH=7.4) buffer solution, and then add to a solution containing 4.0 In the buffer solution of μmol cyanine dye Tris-HCl (pH=7.4), avoid light and stir at 30°C for 0.5h. After the reaction is completed, the reaction solution is added to the dialysis bag for dialysis separation, and then the solution in the dialysis bag is taken out and freeze-dried to obtain Fluorescently labeled polypeptide DGRKLVFFCG. The molecular weight was identified by MALDI-TOF.
[0057] MALDI-TOF: Calculate C 94 h 131 N 16 o 17 S + The m / z is 1787.95, from Figure 7 The m / z obtained in the spectrum shown is 1787.2.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com