Quantitative detection method for specifically detecting mature microRNA expression level and kit
A quantitative detection method and expression level technology, applied in the field of molecular biology, can solve the problems of inability to fully compensate for non-specific amplification of primers, difficult to mature microRNA with high specificity, and expensive Taqman probes.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
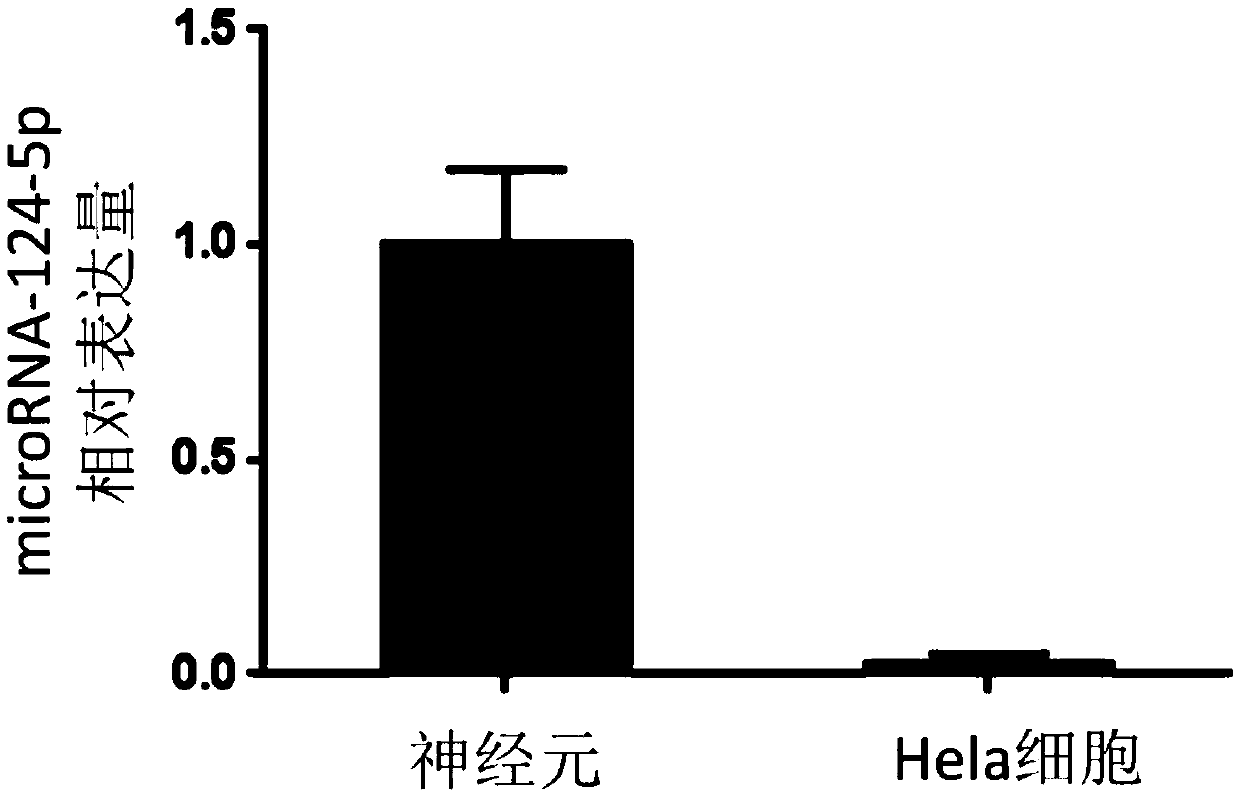
[0048] The expression levels of microRNA-124-5p (MIMAT0004591, miRbase database) in neurons and Hela cells were compared. The specific implementation steps of this test are as follows:
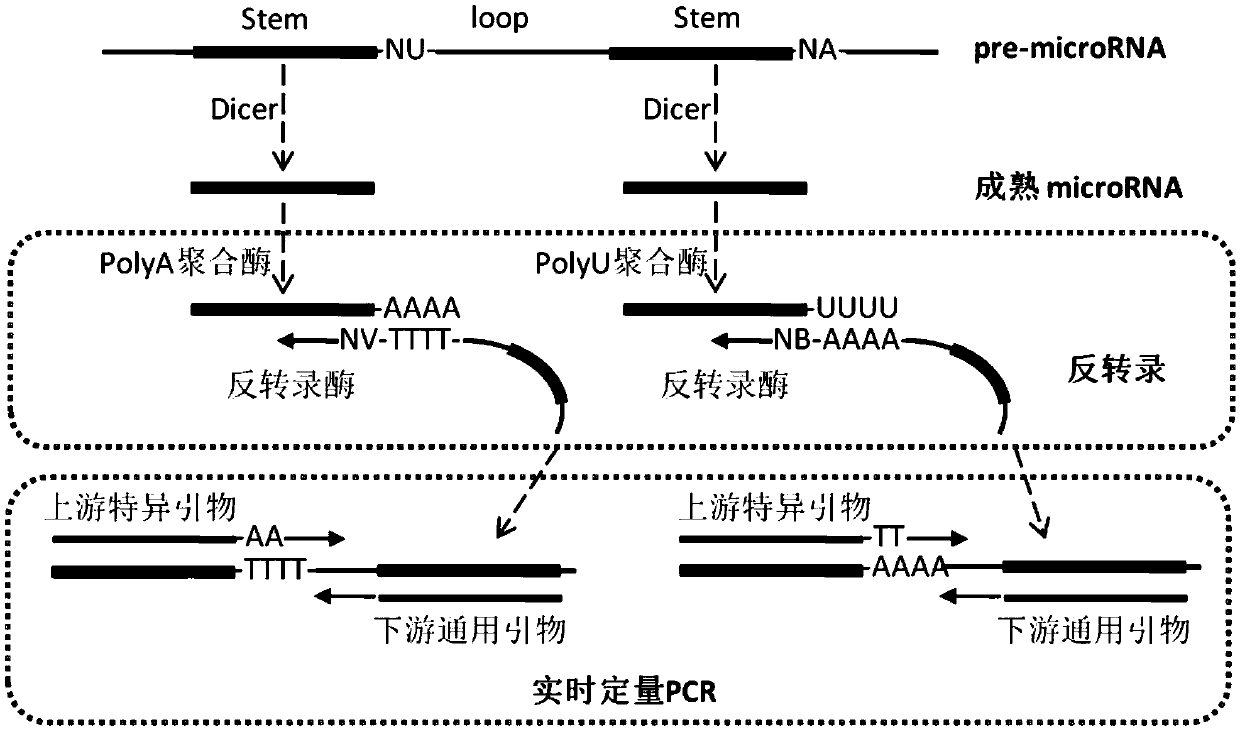
[0049] Reverse transcription process: Use purchased microRNA extraction kits (such as Qiagen, USA) to extract and enrich small RNAs (smallRNAs, including mature microRNAs, pre-microRNAs, sncRNAs, etc.) in neurons and Hela cells. Use purchased (such as U.S. Invitrogen Company) reverse transcription kit, and purchased (such as NEB Company) polyadenosine nucleotide polymerase (poly (A) polymerase) or polyuridine nucleotide polymerase (poly(U) polymerase) mix to reverse small RNAs to cDNA.
[0050] Wherein, the selection mode of polynucleotide polymerase and reverse transcription primer is as follows:
[0051] 1. The sequence of the precursor pre-microRNA of microRNA-124-5p (MI0000443, miRbase database) is as follows:
[0052] AGGCCUCUCUUCUC CGUGUUCACAGCGGACCUUGAU UUAAAUGUCCAUACAAUUAAGGCACGCG...
Embodiment 2
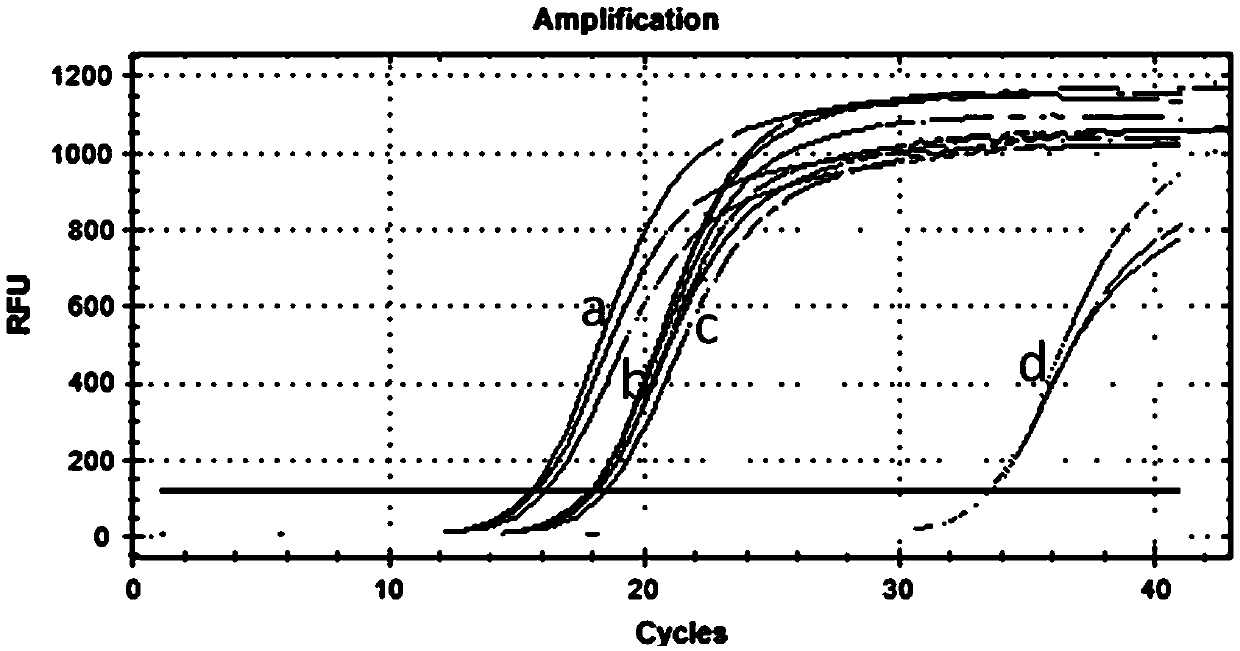
[0081]Comparison of specificities for the amplification of mature microRNAs. While using the method introduced in the present invention to test the expression level of microRNA-124-5p, the currently commonly used Stem-loopPCR method is used as a control. Two methods were used to detect the expression level of microRNA-124-5p in neurons, and the obtained PCR products were electrophoresed simultaneously to test the specificity of the two methods. Use the testing procedure that the present invention introduces as specific embodiment 1. The steps of using stem-loopPCR detection are as follows:
[0082] Reverse transcription process:
[0083] Use the purchased microRNAstem-loop reverse transcription and PCR kit, and reverse-transcribe the enriched small RNA according to the instructions. Concrete reaction system is as shown in table 3:
[0084] table 3
[0085] small RNA
less than 2ug
2x reaction buffer (included in the kit)
10ul
Reverse transcri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com