High throughput test method for SUMO modification substrate in living cells and application thereof
A high-throughput, live-cell technology for protein research that addresses issues such as insufficient sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
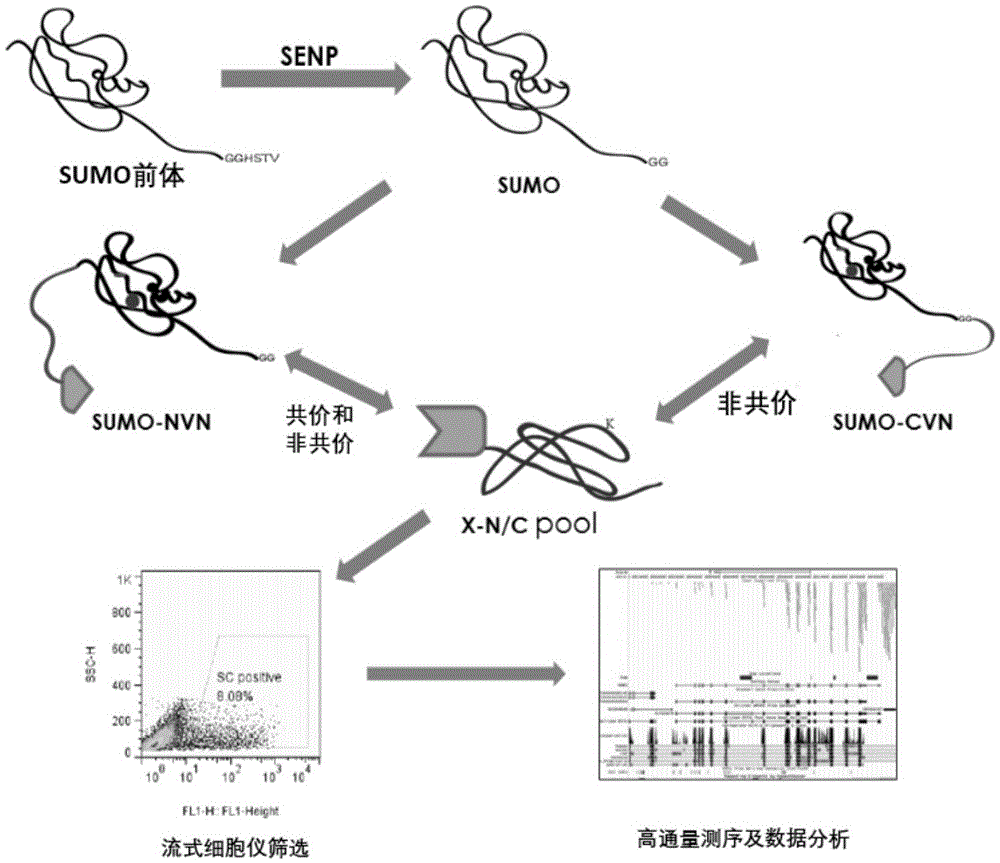
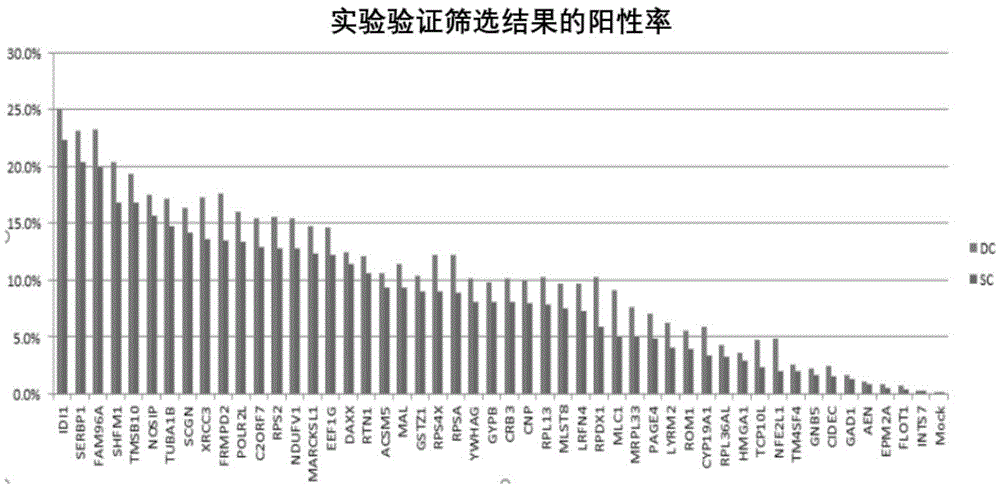
[0067] 1. The method for high-throughput detection of two types of SUMO modified substrates in living cells provided by the present invention is based on the BiFC system, and the reverse transcription expression of the C-terminal and N-terminal links of SUMO1 and SUMO2 / 3 to YFPn is constructed by cloning Vectors (4 libraries), the cDNAs of multiple genes (21000 human genes) of the object to be detected are cloned into the reverse transcription expression vector containing YFPc. After virus infection, the following fusion proteins will be stably expressed simultaneously: SUMO1cvn-YFPn and the gene to be screened-YFPc, SUMO2 / 3C-YFPn and the gene to be screened-YFPc, SUMO1nvn-YFPn and the gene to be screened-YFPc, SUMO2 / 3nvn-YFPn and The gene to be screened-YFPc; if the SUMO substrate interacts with SUMO, observe the fluorescent one at the FITC wavelength by the flow cytometer and detect the signal strength.
[0068] Wherein, the cDNA subcloning of SUMO1, SUMO2 / 3 is fused into th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com