TMEM176A gene promoter region DNA methylation detection
A technology for TMEM176A and gene promoter region, applied in the kit containing the primer pair, non-methylated primer pair, methylated primer pair for detecting the methylation state of TMEM176A gene promoter region, can solve the problem of esophagus Cancer epigenetic changes and functional studies have not yet been reported, and have achieved far-reaching clinical significance and promotion value, good stability, and easy operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] 1. Template preparation (extraction of genomic DNA and sulfuration modification process)
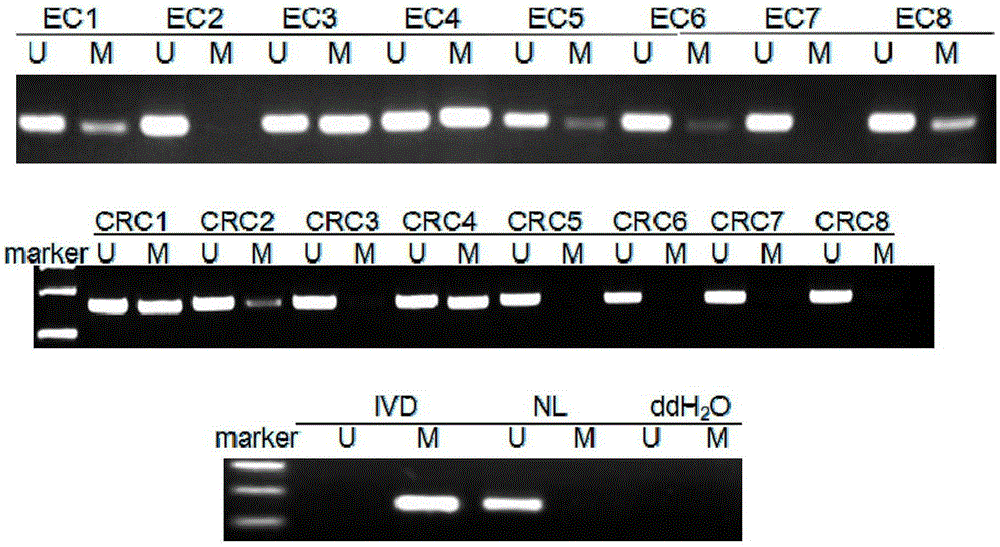
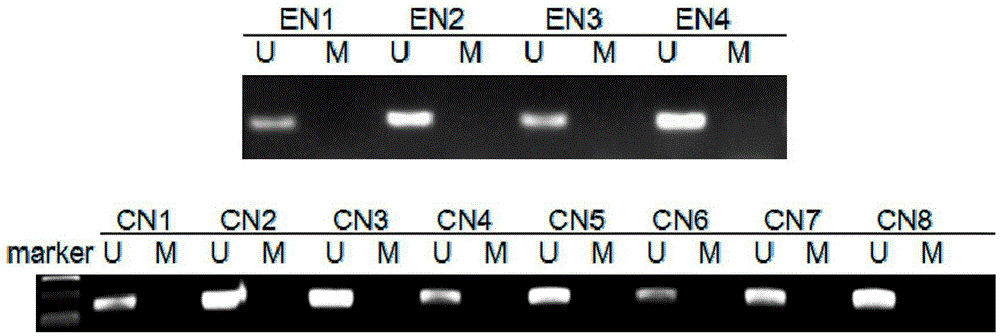
[0044] Preparation of DNA: Obtain esophageal cancer, colorectal cancer specimens and normal tissue specimens mentioned above. In this example, 8 cases of esophageal cancer (EC1-EC8), 8 cases of colorectal cancer (CRC1-CRC8), 4 cases of normal esophagus (EN1-EN4), and 8 cases of normal colorectum (CN1-CN8) were treated with phenol - The method of chloroform extraction was used to extract genomic DNA respectively, and the absorbance (A) value was measured by an ultraviolet spectrophotometer to determine its content and purity.
[0045] Sulfite modification: refer to herman (J.G.Herman, J.R.Graff, S.Myohanen, B.D.NelkinandS.B.Baylin, Methylation-specificPCR: novelPCRassayformethylationstatusofCpGislands, Proc.Natl.Acad.Sci.USA93(1996), 9821–9826.), etc. method of reporting. Take the genomic DNA prepared above, take 2ug DNA accurately after dilution, add deionized water to a final v...
Embodiment 2
[0071] Example 2 Clinical Specimen Detection
[0072] 103 clinical specimens of esophageal cancer, 96 clinical specimens of colorectal cancer, 4 normal esophageal tissue specimens, and 8 normal colorectal tissue specimens were collected. Perform MS-PCR amplification, template preparation, PCR amplification system and conditions, and detection of amplified products are the same as those in Implementation 1. For the detection results, please refer to The following table :
[0073] Classification Number of cases Number of M (methylation) cases Number of cases of U (no methylation) Methylation positive rate Esophageal cancer 103 63 40 61.2% normal esophagus 4 0 4 0 colorectal cancer 96 51 45 53.13% normal colorectal tissue 8 0 8 0
Embodiment 3
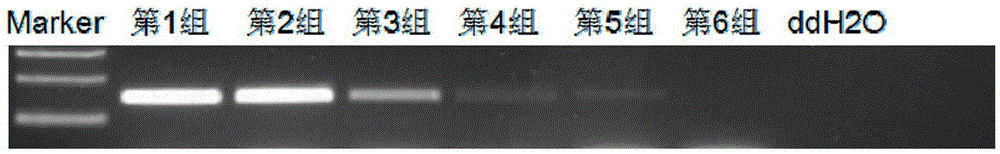
[0074] Embodiment 3 sensitivity experiment
[0075] The DNA of the esophageal cancer cell line K410 (100% methylation in the promoter region of the TMEM176A gene) was mixed with the DNA of normal esophageal tissue cells (100% non-methylation in the promoter region of the TMEM176A gene) in proportion, and the sulfuration modification was carried out (the method was the same as that of Embodiment 1), then carry out MS-PCR. PCR products were subjected to 2% agarose gel electrophoresis, measured by ultraviolet transmission analyzer and photographed.
[0076] Grouping: Group 1: 100% DNA of esophageal cancer cell line K410+0% DNA of normal esophageal tissue cells
[0077] Group 2: 50% DNA of esophageal cancer cell line K410+50% DNA of normal esophageal tissue cells
[0078] Group 3: 5% DNA of esophageal cancer cell line K410+95% DNA of normal esophageal tissue cells
[0079] Group 4: 1% DNA of esophageal cancer cell line K410 + 99% DNA of normal esophageal tissue cells
[0080] ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Upstream primer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com