Method for quantitatively detecting aflatoxin B1
A technology for quantitative detection of aflatoxins, applied in biochemical equipment and methods, measuring devices, instruments, etc., can solve problems such as unfavorable on-site detection, cumbersome operation, complicated pretreatment, etc., and achieve good application prospects and good repeatability. , good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
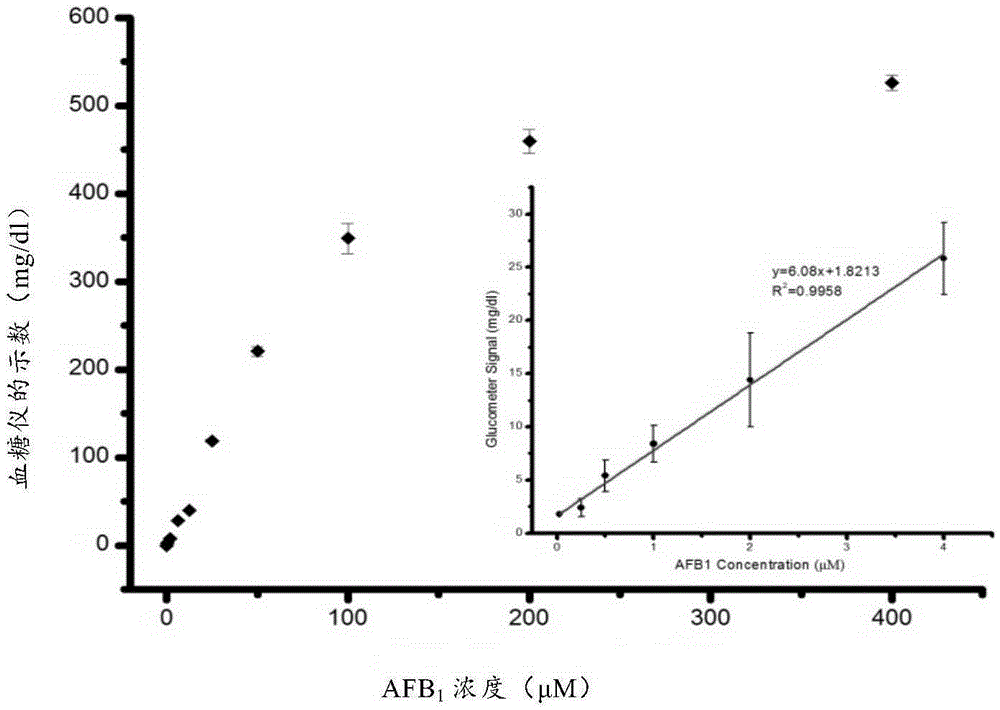
[0046] Example 1 Synthesis of aptamer biosensor and detection of aflatoxin B1 in combination with a blood glucose meter
[0047] 1. Experimental method
[0048] 1.1 Synthesis of DNA-sucrase polymer
[0049] 1.1.1 Activation of sucrase molecules
[0050] Take 400 μl of 20 mg / ml sucrase (buffer B) and mix with 1 mg sulfo-SMCC, vortex and shake for 5 min, place on a constant temperature mixer, and react at room temperature for 2 h.
[0051] 1.1.2 Activation of DNA molecules
[0052] Take 100 μl of 100 μM complementary DNA (thiol-DNA, 3'-SH-A12-CAACCCGTGCACA-5'), 2 μl of 0.1M bufferB, 2 μl of 30mMTCEP (ultrapure water) into a 1.5ml centrifuge tube, vortex to mix, and place in a constant temperature mixer reaction at room temperature for 1 h (among them, ① treatment of complementary DNA: centrifuge the synthesized solid DNA (4°C, 12000 rcf, 5 min), add 345 μl ultrapure water as required, vortex slightly to obtain 345 μl 100 μM DNA solution; ② TCEP Preparation: TCEP molecular we...
Embodiment 2
[0079] Synthesis of Example 2 Aptamer Biosensor
[0080] 1.1 Synthesis of DNA-sucrase polymer
[0081] 1.1.1 Activation of sucrase molecules
[0082] Take 300 μl of 20 mg / ml sucrase (buffer B) and mix with 0.5 mg sulfo-SMCC, vortex and shake for 5 min, place on a constant temperature mixer, and react at room temperature for 1 h.
[0083] 1.1.2 Activation of DNA molecules
[0084] Take 80μl of 100μM complementary DNA (thiol-DNA, 3'-SH-A12-CAACCCGTGCACA-5'), 1μl of 0.1MbufferB, 1μl of 30mMTCEP (ultrapure water) and add it to a 1.5ml centrifuge tube, vortex and mix well, and place in a thermostatic mixer On, react at room temperature for 0.5h.
[0085] 1.1.3 Synthesis of DNA-sucrase polymer
[0086] Centrifuge the reaction solution of sucrase-SMCC and thiol-DNA (25°C, 12000rcf, 5min), absorb the supernatant, and add them to ultrafiltration tubes (Amicon-100K for sucrase-SMCC; Amicon-100K for thiol-DNA) 3K), centrifuge (25°C, 12000rcf, 10min), wash 8 times with bufferA; suck ...
Embodiment 3
[0094] Example 3 Synthesis of Aptamer Biosensors
[0095] 1.1 Synthesis of DNA-sucrase polymer
[0096] 1.1.1 Activation of sucrase molecules
[0097] Mix 500 μl of 20 mg / ml sucrase (buffer B) with 2 mg of sulfo-SMCC, vortex for 5 min, place on a constant temperature mixer, and react at room temperature for 3 h.
[0098] 1.1.2 Activation of DNA molecules
[0099] Take 120 μl of 100 μM complementary DNA (thiol-DNA, 3'-SH-A12-CAACCCGTGCACA-5'), 3 μl of 0.1M bufferB, 3 μl of 30mMTCEP (ultrapure water) and add it to a 1.5ml centrifuge tube, vortex and mix well, and place in a constant temperature mixer On, room temperature reaction 2h.
[0100] 1.1.3 Synthesis of DNA-sucrase polymer
[0101] Centrifuge the reaction solution of sucrase-SMCC and thiol-DNA (25°C, 12000rcf, 5min), absorb the supernatant, and add them to ultrafiltration tubes (Amicon-100K for sucrase-SMCC; Amicon-100K for thiol-DNA) 3K), centrifuge (25°C, 12000rcf, 10min), wash 8 times with bufferA; suck out the thi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com