Oligonucleotide linker and applications of oligonucleotide linker in construction of nucleic acid sequencing single-chain cyclic library
A technology of oligonucleotides and sequencing libraries, applied in the field of oligonucleotide adapters and using the adapters to construct nucleic acid sequencing single-stranded circular libraries, which can solve the problem of affecting experimental efficiency and the construction of unsatisfactory single-stranded circular libraries method, increasing operational complexity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0135] Example 1 Construction of RNA single-stranded circular library
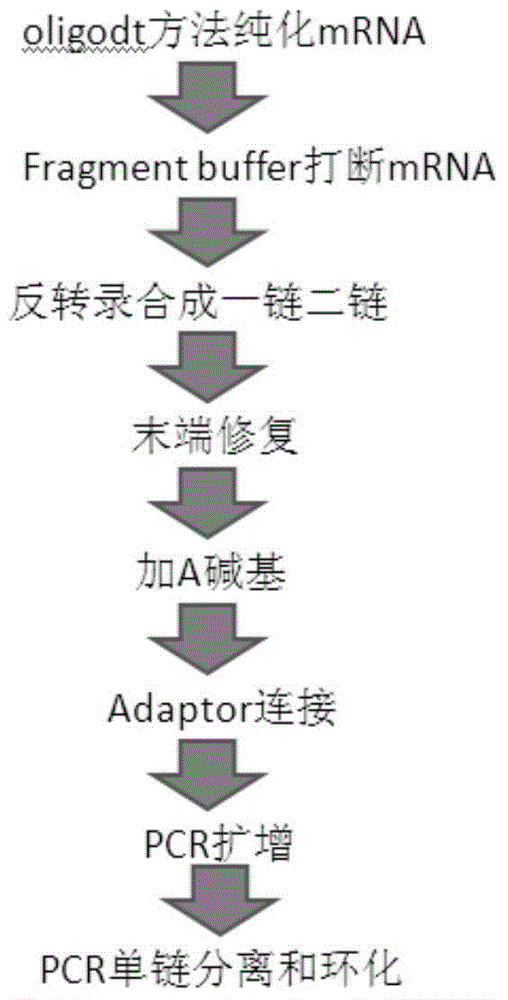
[0136] Specific experimental steps (see figure 1 Process steps shown in ):
[0137] 1. mRNA extraction and purification
[0138] 1) Take the standard universalhumanreferenceRNA3ug to an RNase-free tube, dilute to 50μl with DEPC water. Mix well, denature at 65°C for 5 minutes to open up secondary structures, and immediately place samples on ice.
[0139] 2) Draw 15μl DynalbeadsOligo(dT) 25 Put the magnetic beads in a 1.5ml non-stick EP tube, wash the magnetic beads twice with 100 μl binding buffer, resuspend the magnetic beads in 50 μl binding buffer, add the total RNA prepared in the first step to the tube, and keep at room temperature Leave it for 5min.
[0140] 3) Place the non-stick-EP tube on the MPC (magnetic separator) for 2 minutes, remove the supernatant, and wash the magnetic beads twice with 200 μl washing buffer. Take a new non-stick EP tube and add 50 μl of binding buffer (bindingbuffer)....
Embodiment 2
[0222] The library preparation steps 1-10 in Example 1 were repeated, except that conventional human lymphocytes (YHcell) were used instead of the standard universalhuman reference RNA in Example 1 as the source of total RNA. Use the QIAGEN RNA extraction kit to extract the total RNA of Yanhuang cells. Before the extraction and purification of mRNA, the total RNA is pretreated. The pretreatment steps are as follows:
[0223] 1. DNase digestion of total RNA
[0224] Take the total RNA sample, put it in an RNase-free EP tube, and carry out DNA digestion according to the following system.
[0225]
[0226] Digest at 37°C for 10 minutes.
[0227] Add 2μl 1mM EDTA solution, denature at 75°C for 10min, and cool on ice.
[0228] 2. Purification of digested RNA
[0229] Add the product (22 μl) of the above steps to 1.8 times the volume (40 μl) of RNAclean magnetic beads (need to equilibrate at room temperature for 30 minutes in advance). Place at room temperature for 5 minutes,...
Embodiment 3
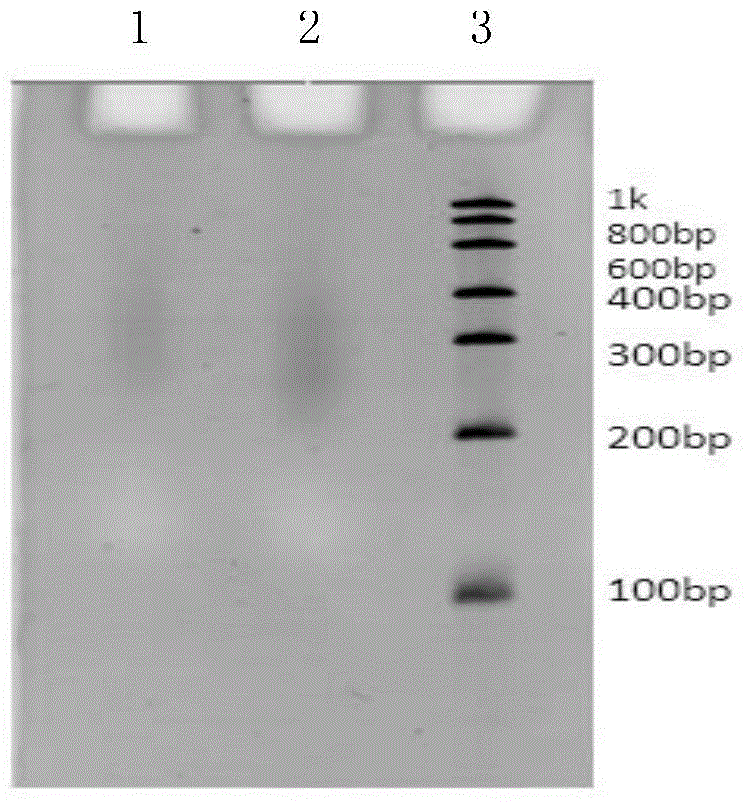
[0242] The DNA of Hela tumor cells was extracted with QIAGEN High Quality DNA Extraction Kit. Then repeat steps 4 to 9 in Example 1, 6% TBE denatured gel detection.
[0243] see results Figure 7 , the results show that it is feasible to use the method of the present invention to prepare a DNA sequencing library.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com