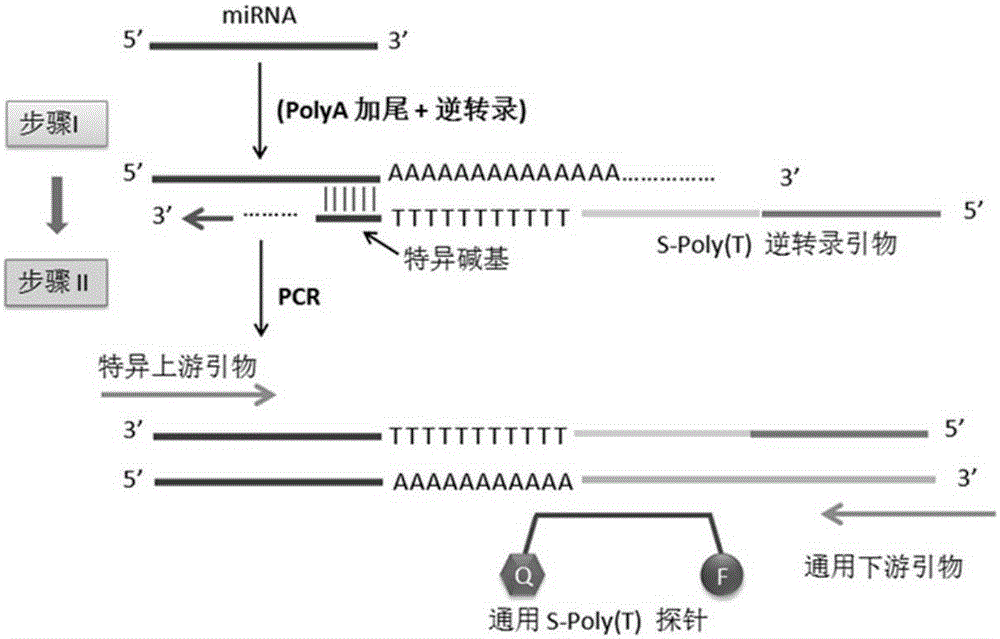
RT-PCR method for quantitatively detecting miRNA
A RT-PCR, quantitative detection technology, applied in the field of biomedicine, can solve problems to be improved and improved, and achieve the effects of simple operation, improved sensitivity and accuracy, and excellent simplicity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Example 1, S-Poly(T)Plus method for detection of circulating miRNA
[0059] The S-Poly(T)Plus method for detecting circulating miRNA is as follows: Figure 13 shown, including the following steps:
[0060] (1), extraction of serum total RNA
[0061] In this embodiment, the S / PmiRsol method is used to extract serum total RNA, and the specific steps are:
[0062] 1) Add 0.1pM nematode miRNAcel-miR-54 as an internal reference to 1mL RNAiso-Plus (TaKaRa) in advance, add 100μL serum, pipette and mix, let stand at room temperature for 5min; add 200μL chloroform, tightly cap the centrifuge tube, and shake vigorously for 20s ;Stand at room temperature for 5min;
[0063] 2) Centrifuge at 12,000g at 4°C for 15min; take out the centrifuge tube carefully, and the homogenate is divided into three layers, namely: a colorless supernatant (containing miRNA), a white protein layer in the middle, and a colored lower organic phase ;Pipe 500μL supernatant and transfer to another new ...
Embodiment 2
[0096] Example 2, S-Poly(T)Plus method for detecting miRNA in cells
[0097] (1) Extraction of total RNA in cells
[0098] In this embodiment, the specific steps for extracting total cellular RNA are:
[0099] 1) Add 10 6 For individual 293A cells, pipette and mix well, and let stand at room temperature for 5 minutes; add 200 μL of chloroform, tightly cap the centrifuge tube, shake vigorously for 20 seconds; let stand at room temperature for 5 minutes;
[0100] 2) Centrifuge at 12,000g at 4°C for 15min; take out the centrifuge tube carefully, and the homogenate is divided into three layers, namely: a colorless supernatant (containing miRNA), a white protein layer in the middle, and a colored lower organic phase ;Pipe 500μL supernatant and transfer to another new 1.5mL centrifuge tube;
[0101] 3) Add an equal volume of isopropanol (500 μL) to the supernatant, mix well by inverting up and down, and let stand at -20°C or -80°C for at least 10 minutes;
[0102] 4) Centrifug...
Embodiment 3
[0129] Example 3, S-Poly(T)Plus method detects the linear gradient range of miRNA in cells
[0130] This example analyzes the linear gradient range of the S-Poly(T)Plus method for detecting miRNA in cells. The human 293 cell RNA was diluted 5 times (2.5ng~0.8pg), and then six miRNAs (miR-92a-3p, miR-16-5p, miR-27b-3p, miR-210-3p, miR -103a-3p and miR-126-3p) and two internal reference snoRNAs (SNORD44 and SNORD7). From Figure 5 It can be seen from the figure that the linear correlation coefficient R of S-Poly(T)Plus detection of different miRNAs 2 (0.9933~0.9991) are greater than the value (0.9745~0.9968) of corresponding miRNA detected by S-Poly(T) method. Therefore, the S-Poly(T)Plus method for detecting cellular miRNA has a good linear relationship and a wide dynamic range.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com