Method for high yield production of aerobic single-cell protein by autolysis process
A single-cell protein, high-yield technology, applied in the direction of microbial/single-cell algae protein processing, etc., can solve the problems of not considering the decomposition of nucleic acid into high-value nucleotides, failing to reach the average production capacity of processing equipment, and low protein digestibility , to achieve the effect of controllable residence time, prevention of diarrhea, and rapid heating
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
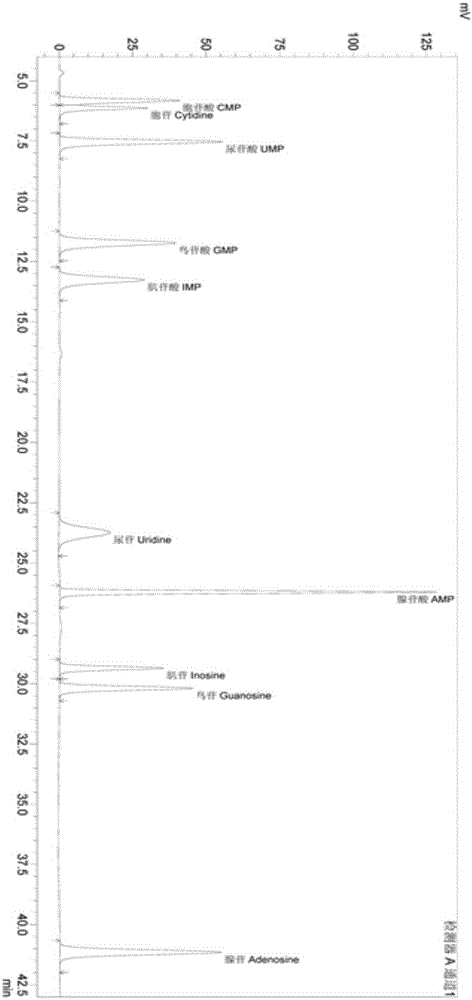
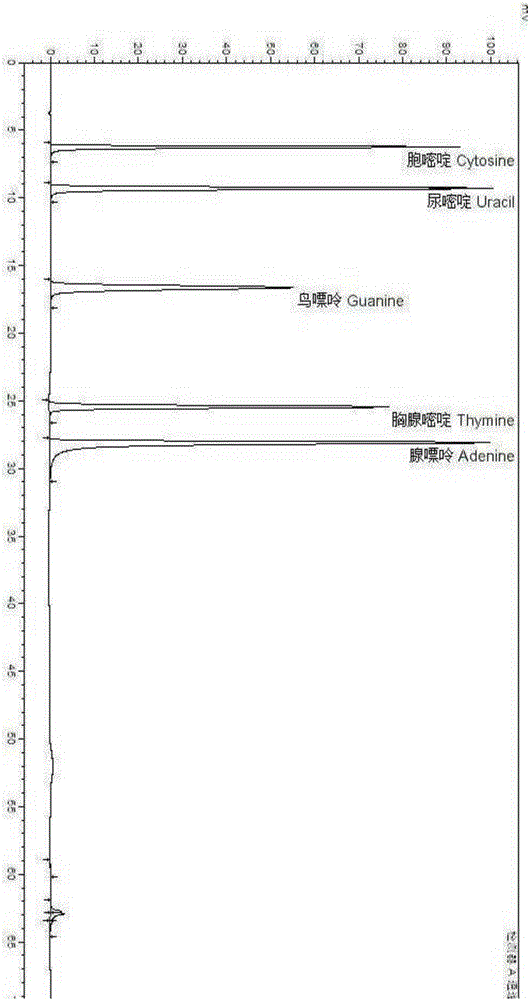
[0053] Embodiment 1: the establishment of free nucleotide and free base detection method
[0054] Use free nucleotides and free bases as the main indicators for the adjustment of autolysis process parameters and the comparison of products before and after autolysis. The principle is: raw materials such as yeast and bacterial protein will contain free nucleotides and bases after autolysis or enzymatic hydrolysis, and the sample extracted from the aqueous solution will be analyzed by high-performance liquid chromatography. Salt is used as the mobile phase, and the external standard method can be used for quantitative determination of 5 nucleotides, 5 nucleosides and 5 bases.
[0055] Free nucleotides refer to the sum of 5 nucleotides and 5 nucleosides, including cytidylic acid, uridine acid, guanylic acid, inosinic acid, adenylic acid, cytidine, uridine, inosine, guanylic acid glycosides, adenosine. Free base refers to the sum of 5 nucleic acid bases, including cytosine, uraci...
Embodiment 2
[0119] Embodiment 2: autolysis production technology and product
[0120] The autolysis production process and products developed by the present invention are single-cell protein products made by using the cultivation technology of the invention patent WO2009059163A1, but are not limited thereto, and are also applicable to the processing of aerobic single-cell protein products similar to this invention.
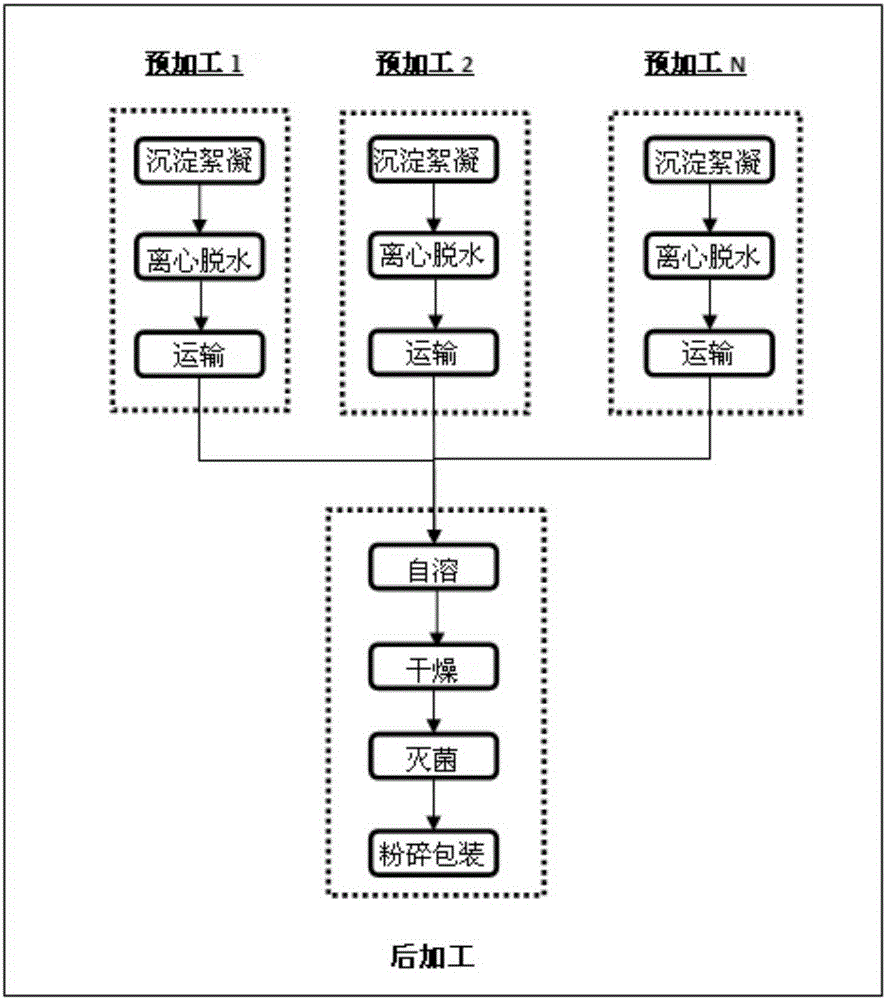
[0121] A high-yield processing method of autolyzed single-cell protein is realized by dividing the processing process into two parts, which are respectively completed by different production units. The first part is the pre-processing unit, which includes the process from precipitation concentration to centrifugal dehydration after the single-cell protein culture is completed. In this part, multiple preprocessing units can be set, for example, 2 to 10 units are all possible. The second part is the post-processing unit, which includes the operations of autolysis, drying and s...
Embodiment 3
[0162] Embodiment 3: digestibility test
[0163] The difference in apparent digestibility between the bacterial protein using the autolysis process and the common bacterial protein was evaluated by animal experiments.
[0164] Digestibility is the percentage of digestible nutrients in the feed ingredients ingested by animals. The indicator method is to use the indicator existing in the feed or artificially mixed evenly. After feeding the fish, the digestibility was calculated from the amount of the unit indicator contained in the unit feed and feces. Formulating formulas based on the digestible value of nutrients in feed ingredients is of great significance for improving the digestibility of compound feed and reducing the pollution of feed substances to the aquaculture water environment.
[0165] Test plan:
[0166] Raw materials for evaluation: The bacterial protein with a protein content of 50% was autolyzed according to the method of Example 2, and the same batch of sing...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com