Saccharomyces cerevisiae engineering strain as well as preparation method, application and fermentation culture method thereof
A technology for Saccharomyces cerevisiae and engineering strains, which is applied in the field of bioengineering to achieve the effect of simple medium components
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] Construction of Example 1 Deletion Product
[0084] 1. Synthetic exogenous gene
[0085] According to the sequence of lactate dehydrogenase gene NM_174099, its DNA was artificially synthesized after optimization by J-Cat.
[0086] 2. Build missing modules
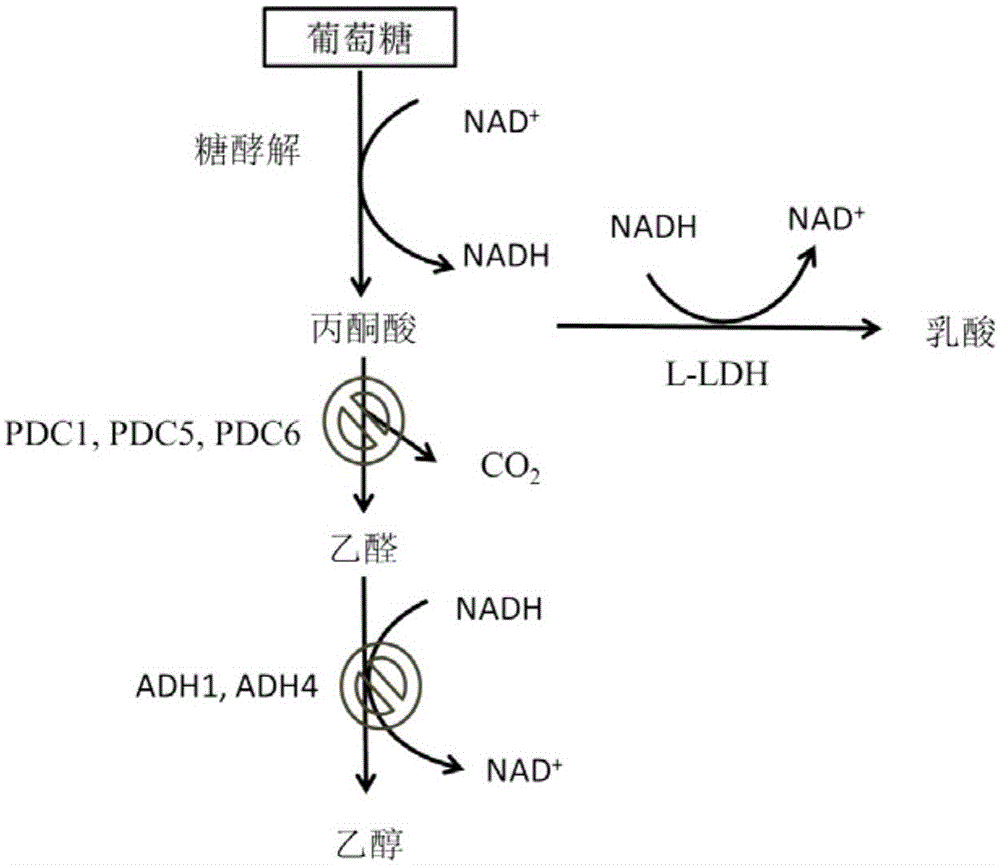
[0087] Using the genome of Saccharomyces cerevisiae BY4741 as a template, the promoters, terminators and ORF boxes of pyruvate decarboxylase genes PDC1, PDC5 and PDC6 and acetaldehyde dehydrogenase genes ADH1 and ADH4 were amplified. The amplification primer sequences of each part are as follows:
[0088] Table 1 Amplification Primer Sequence
[0089]
[0090]
[0091]
[0092] According to the PDC5-related primer sequences in Table 1, the promoter, terminator, nutritional marker URA3 and ORF box part of the pyruvate decarboxylase gene PDC5 were amplified, and the above products were connected together by PCR reaction to obtain the deletion module ΔPDC5 as figure 2 shown.
[0093] The promoter, termina...
Embodiment 2
[0097] Embodiment 2: Construction of gene knockout strain
[0098] 1. Inoculate BY4741 strain in YPD medium and culture overnight at 30°C.
[0099] 2. Transfer the culture solution of the above overnight strain to 5ml of fresh YPD to make the initial OD 600 =0.1, continue culturing at 30°C for 4-6 hours to make the culture medium OD 600 Reach 0.4-0.6.
[0100] 3. Transformation: The deletion module ΔPDC5 obtained above was transformed into competent cells made of BY4741 by the lithium acetate transformation method, and then spread on SC-Ura plates after transformation and cultured at 30°C.
[0101] 4. Genome extraction verification: pick the transformant, extract the genome for PCR verification label, BY4741 is used as a negative control, and the band size is correct, indicating that the homologous recombination of the missing module is correct and completed.
[0102] 5. Pop off the nutritional marker Ura3: the correct transformant will be verified and marked on FoA, and th...
Embodiment 3
[0120] Example 3 Molecular biology verification
[0121] The extraction method of the yeast genome refers to the yeast genetics experiment guide, and the primers used in the molecular biology verification of Example 2 are as follows:
[0122] 1. After transforming the missing module, the PCR verification primers for Ura3 were not removed:
[0123] The primers for the verification of deletion module ΔPDC5 recombination were upstream p5_cku: 5'-TTTCAGCTCTTTCAAGTTCCTC-3' (SEQ ID NO: 45), downstream P_p5_dw: 5'-CATGAGTTTTATGTTAATTAGCTTATTTGTTCTTCTTGTTATTGTATTGTGTTGTTC-3' (SEQ ID NO: 46), and upstream U_p5_ACT: 5'-TCCTTAAGAT3ATGCTCGAATACTAAGAG (SEQ ID NO: 47), downstream p5_ckd1: 5'-TAGACTGGTCTTTGGGTAGTGTAGG-3' (SEQ ID NO: 48).
[0124] The primers for the verification of deletion module ΔPDC6 recombination were upstream p6_cku: 5'-ATGTCCATTGGAATATGCAGA-3' (SEQ ID NO: 49), downstream P_p6_dw: 5'-GTTTGAGTACACTACTAATGGCTTATTTGTTGGCAATATGTTTTTGCTATATTACGTG-3' (SEQ ID NO: 50), and ups...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com