A method for preparing multiple DNA large fragments inserted into a paired-end sequencing library at one time
A paired-end sequencing and large-fragment technology, applied in the field of molecular biology, can solve the problems of multiple preparations and waste of DNA samples, and achieve the effects of reducing the cost of building a library, avoiding waste, and reducing demand
Active Publication Date: 2017-09-05
深圳中农京跃生物技术有限公司
View PDF7 Cites 0 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
[0004] The purpose of the present invention is to provide a method for preparing multiple large DNA fragments inserted into a paired-end sequencing library at one time, aiming at solving the following defects in the prior art: first, only large fragment paired ends containing one insert length can be prepared at a time Sequencing libraries, in practical applications, when large fragment libraries with different insertion lengths are required, multiple preparations are required; second, when the same DNA sample is used to construct large fragment libraries with different insertion lengths, often only one target fragment (only Accounting for 10-20% of the total DNA), resulting in the waste of a large number of DNA samples
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment 1
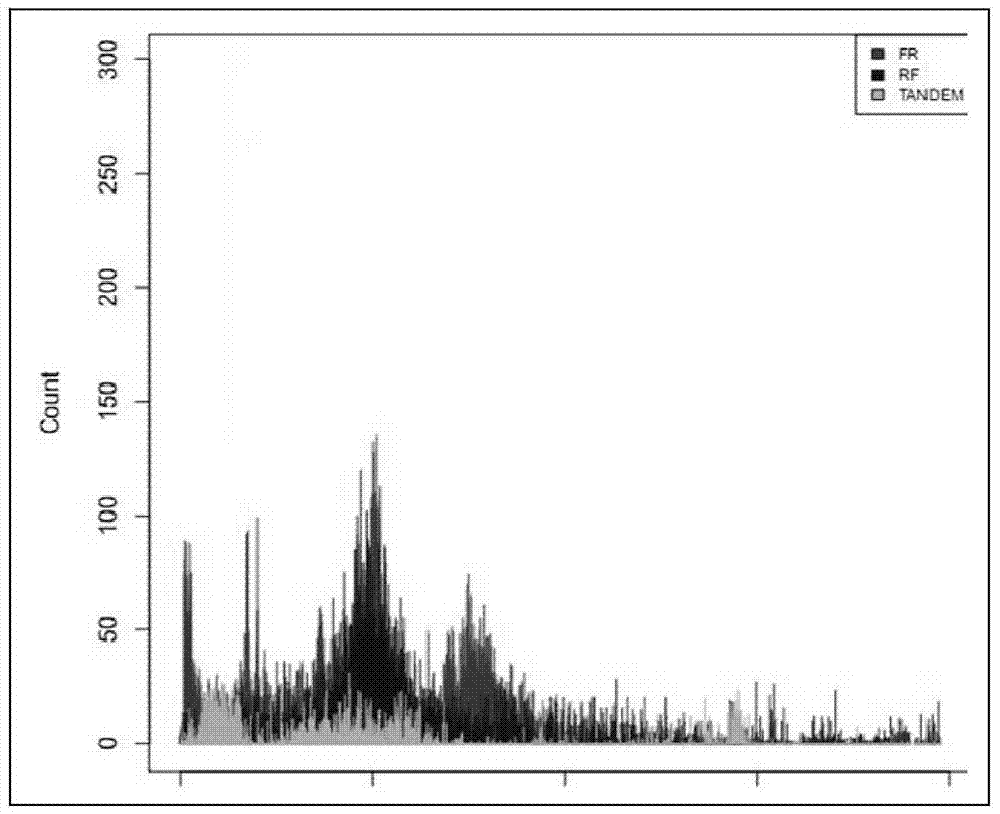
[0086] attached figure 2 The peak distribution diagram of the large fragment library data containing 11 fragments prepared for the embodiment of the present invention is obtained by figure 2 It can be seen that the span of each fragment is very narrow, which is very beneficial for subsequent genome assembly and structural variation analysis.
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
The invention provides a method for preparing a plurality of large DNA fragments to be inserted into a paired-end sequencing library at one time, comprising the following steps: performing biotin linker labeling after the DNA to be tested is broken; sorting the biotin-labeled DNA fragments into N DNA fragments of different sizes, wherein, 10≤N≤12; the N DNA fragments of different sizes are respectively subjected to circularization treatment to obtain circular DNA, and then the uncircularized linear DNA fragments are removed, and the circular DNA After DNA fragmentation, connect Truseq adapters with different index sequences; mix DNA fragments of different lengths with Truseq adapters and react with streptavidin magnetic beads to enrich biotin-labeled DNA fragments; PCR The biotinylated DNA fragments are amplified and sequenced.
Description
technical field [0001] The invention belongs to the field of molecular biology, and in particular relates to a method for preparing a plurality of large DNA fragments inserted into a paired-end sequencing library (N-Size-Fragment Mate Pair Library, NSF-MP Library). Background technique [0002] Large fragment insertion library (Mate-paired library) plays an important role in genome research. In genome de novo sequencing, large fragment insertion library can provide a skeleton for genome assembly and improve the accuracy and quality of genome assembly. The currently completed large whole genome de novo sequencing is composed of several small fragment libraries (shotgun library) and large fragment libraries (2kb, 5kb, 10kb and 20kb) with inserts of different sizes (such as 200bp, 400bp). Use the reads from the small fragment library to assemble the contig, and then use the reads from the large fragment library to assemble the contig into a scaffold. In addition, large fragmen...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Patents(China)
IPC IPC(8): C40B50/06C12Q1/68
Inventor 常玉晓赵胜刘可
Owner 深圳中农京跃生物技术有限公司
Who we serve
- R&D Engineer
- R&D Manager
- IP Professional
Why Patsnap Eureka
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com