Error-prone whole-genome shuffling approach for Escherichia coli and butanol-tolerant Escherichia coli
An Escherichia coli and tolerance technology, applied in the field of bioengineering, can solve the problems of operator hazards, time-consuming and labor-intensive adaptation to evolution, etc., and achieve the effects of safe and environmentally friendly reagents, fast and efficient operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Genomic DNA Extraction:
[0034] The genomic DNA of the starting strain Escherichia coli BW25113 was extracted, and the specific operation was as follows: the above bacterial solution in the glycerol tube was transferred to a test tube containing 5 mL of LB liquid medium, and cultured overnight at 37°C to make OD600=2-3. Transfer the culture to a centrifuge tube, centrifuge at 13,000 rpm at 4°C for 1 minute, discard the supernatant, resuspend the pellet with 30 μL of sterile water, and discard the supernatant by centrifugation. Add 120 μL of bacteriostasis buffer, 60 μL of Tris saturated phenol, 60 μL of chloroform, 120 μL of TE buffer, and 120 μL of quartz sand into the centrifuge tube. After fully shaking for 4 min, centrifuge at 13000 rpm / 5 min at 4 °C to obtain the supernatant. Add 1mL of absolute ethanol to the supernatant, mix it upside down, place it at -20°C for 30min, centrifuge at 13000rpm / 25min at 4°C, discard the supernatant, dry the precipitate, a...
Embodiment 2
[0040] Example 2 Error-prone PCR amplification of the whole genome:
[0041] Take 5 μL of dNTPS stock solution, 16.6 μL of 100 uM 10-17 base random primers, 3 μL of 15 mM MnCl 2 , 5 μL of 10× mutation buffer, 20ng of genomic DNA obtained in step (1), 2.5 μL of 2U / μL Taq DNA polymerase, supplemented with sterile water to 50 μL; the dNTPS stock solution contains 10mM dCTP and dTTP, 2mM dATP and dGTP ; The 10×mutation buffer contains 1mM Tris‐HCl pH8.3, 0.7mM MgCl 2 , 5mM KCl, 1g / 100mL glycerol;
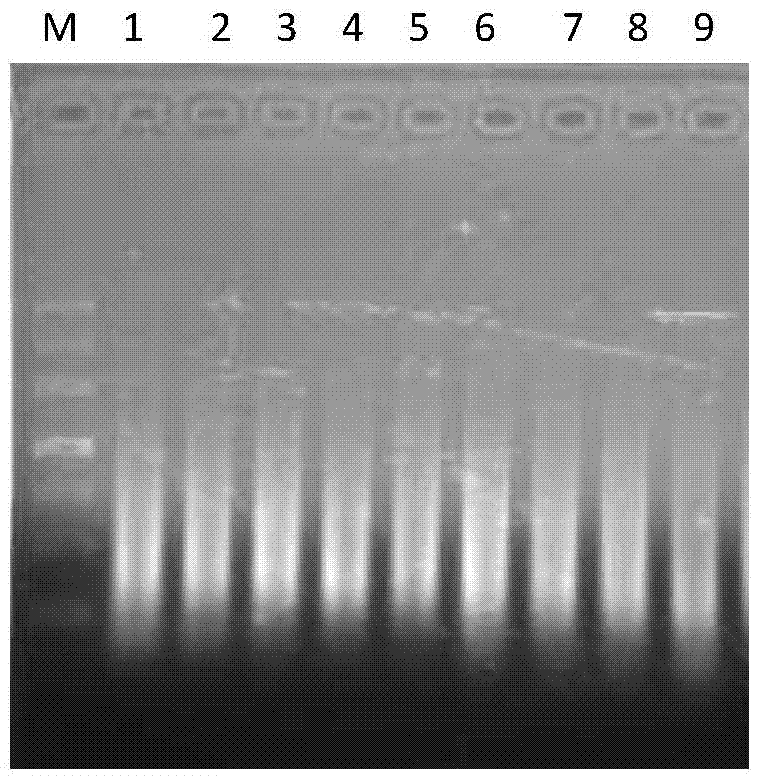
[0042] Error-prone PCR reaction conditions: 94°C pre-denaturation for 5 minutes; 92°C for 1 minute, then annealing at 37°C for 1 minute, then rising from 37°C to 55°C at a rate of 0.1°C per second, and extending at 55°C for 4 minutes. 50 cycles, supplementary extension at 55°C for 10 min; amplification products such as figure 1 shown.
[0043] 16.6μL of 100uM any primer with 10-17 bases can amplify error-prone PCR products.
[0044] 16.6μL of 100uM10-17 bases are:
[0045] 8.3 μL ...
Embodiment 3
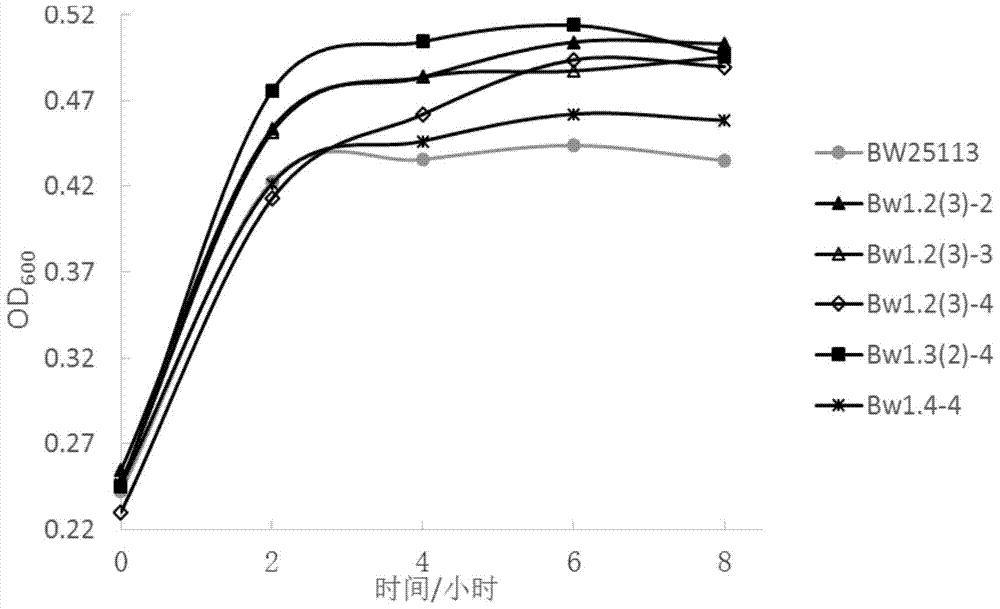
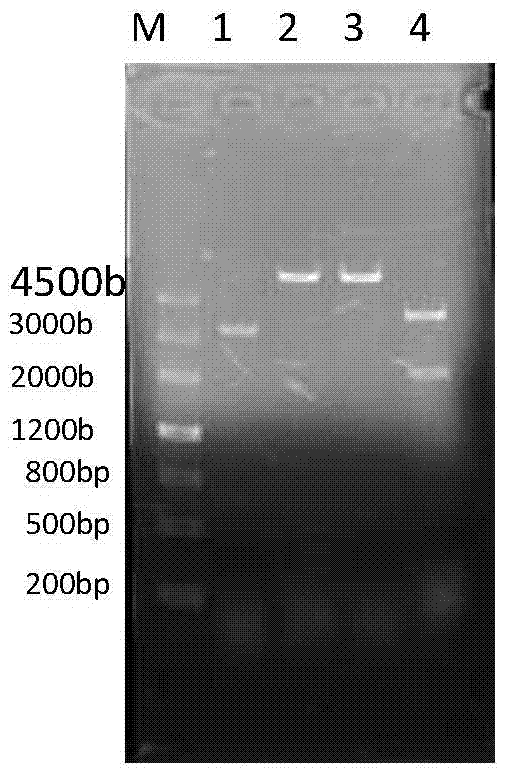
[0065] Example 3 Whole genome shuffling:
[0066] The various PCR products obtained in Example 2 were mixed and concentrated by ethanol precipitation 10 times for electrotransformation;
[0067] Inoculate Escherichia coli BW25113 containing the pKD46 plasmid into 3 mL of LB liquid medium containing 50 ug / mL ampicillin, and culture overnight at 30°C; the next day, transfer it to 50 mL of LB liquid containing 50 ug / mL ampicillin at a ratio of 1% culture medium at 30°C until OD 600 =0.15~0.2, add L-arabinose to a final concentration of 20mmol / L, induce plasmid pKD46 to express Red recombinant protein; cultivate to OD 600 =0.45~0.5, take it out and pre-cool on ice for 10-15min, centrifuge at 5000g at 4°C for 10min, discard the supernatant, wash the bacteria once with 20mL of ice-cold sterile water, and wash twice with 20mL of pre-cooled 10% glycerin. Once concentrated 100 times, resuspend and mix with 500uL ice-cold 10% glycerol to obtain competent cells.
[0068] Take 4 parts ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com