Amplimers, kit and method for detecting PKD1 gene mutation
A kit and gene technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of cumbersome methods, too many primer sets, short read sequences, etc., and avoid pseudogene interference , simplification of experimental operation, and the effect of small GC bias
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Genomic DNA Extraction
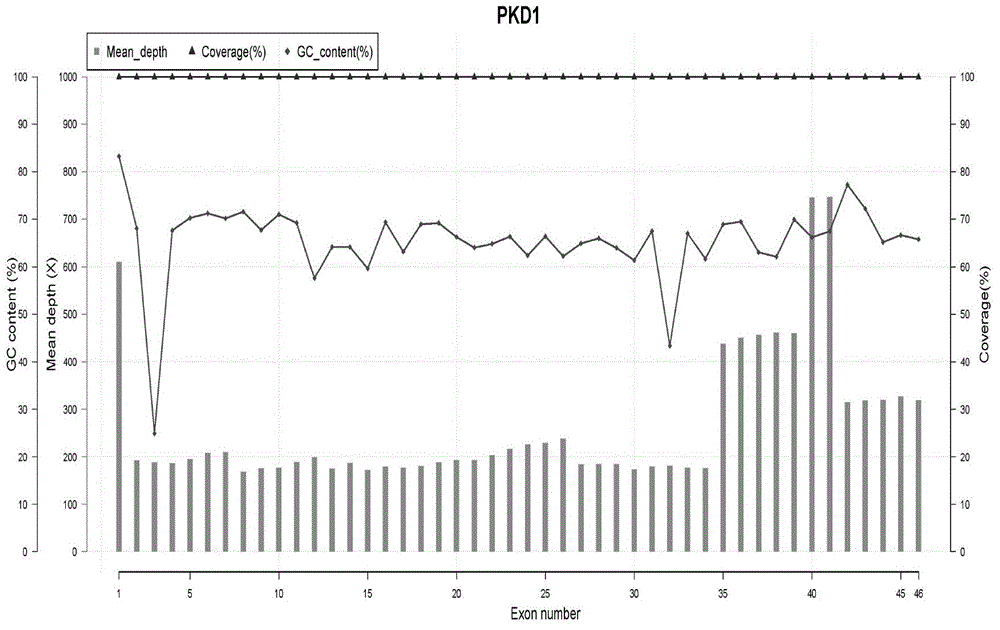
[0033] Collect 2ml of peripheral blood from the subject and place it in an EDTA anticoagulant tube. Take 0.2 ml of EDTA anticoagulated peripheral blood sample, and extract DNA according to the instructions of the whole blood DNA extraction kit. The DNA concentration and purity were analyzed by Qubit 2.0, and the extracted genomic DNA was ready to be used as the next PCR template. In this study, a total of 6 gene DNA samples were detected, of which 4 gene DNA samples had known pathogenic mutation sites, and 2 were normal human genomic DNA.
Embodiment 2
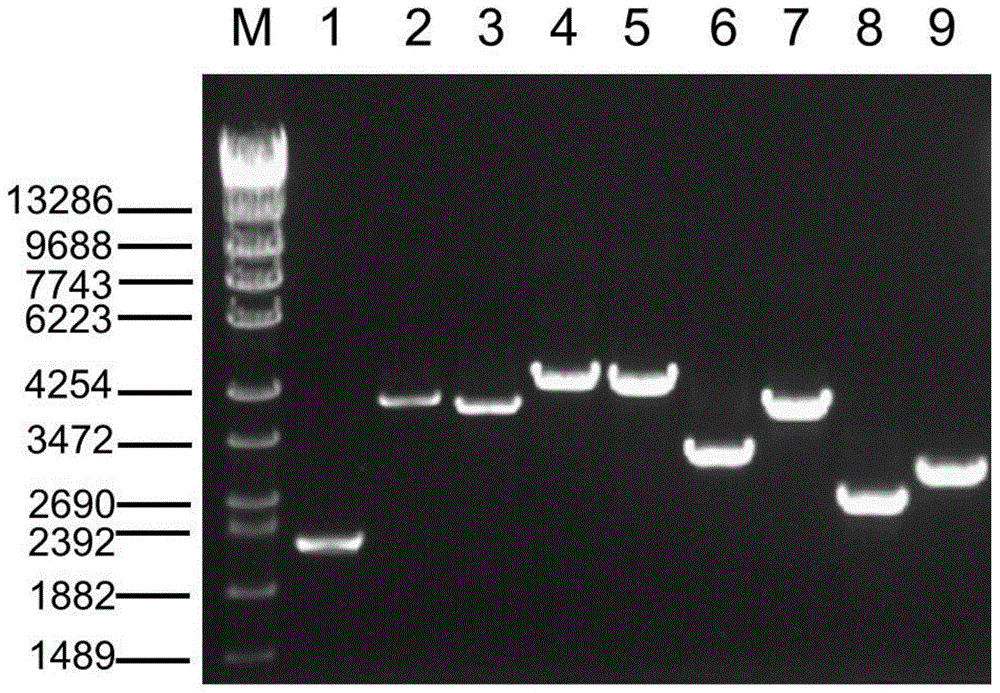
[0035] LR-PCR amplification
[0036] According to the PKD1 gene sequence and its homologous pseudogene sequence, 9 pairs of PKD1 gene-specific primers were designed and synthesized, and GeneAmp High Fidelity PCR System was used for LR-PCR to amplify 9 large fragment sequences, respectively 1, 2, 3, 4, 5, 6, 7, 8 and 9, the primer sequences and amplified product sizes are shown in Table 1. The GeneAmp High FidelityPCR System was used, and the reaction conditions were set to perform LR-PCR on a Veriti 96-well Thermal Cycler PCR instrument. The total volume of the PCR reaction is 50 μl, including 5 μl of 10×GeneAmp High Fidelity PCR buffer, 2 μl of 10 mmol / L dNTP, 1 μl of 10 μmol / L upstream and downstream primers, 10 μl of 5 mol / L betaine, and 2.5 μl of DMSO (for the amplification of fragment 1 Use 5 μl), 1 μl of 5U / μl polymerase mixture, 5 μl of 20 μg / μl DNA template, and make up to 50 μl with ultrapure water. PCR cycle parameters: deformation at 94°C for 3min, 94°C for 10sec,...
Embodiment 3
[0043] LR-PCR Amplification Based on Primer Index Technology
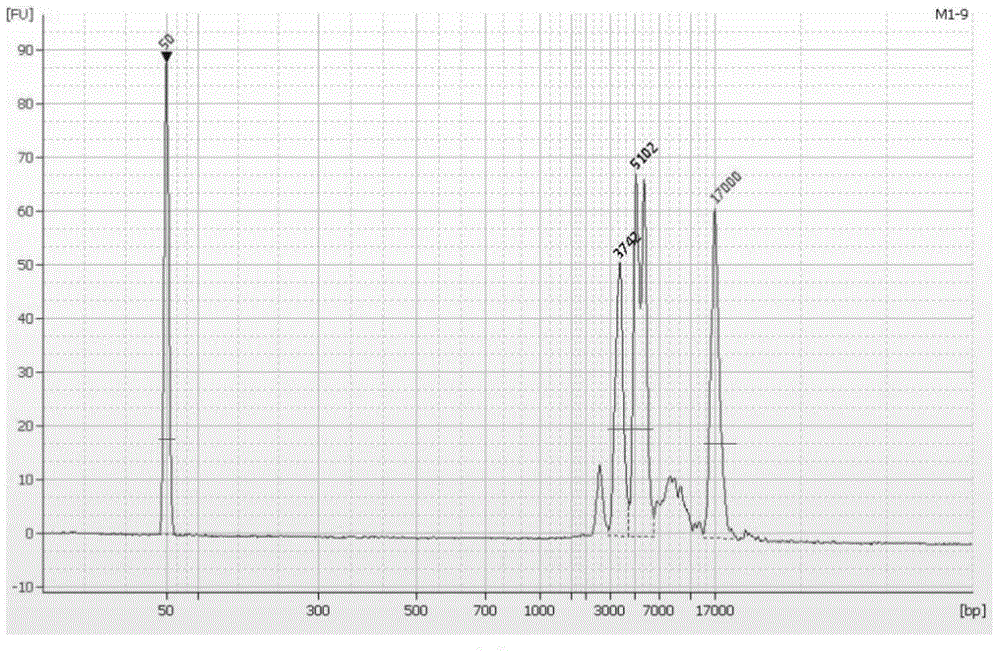
[0044] According to PacBio's Guidelines for Using PacBio Barcodes for SMRT Sequencing, asymmetric label primers were designed on the basis of the above Table 1. The label sequence was referred to the 16-base label (Barcode) sequence provided by PacBio, and the interaction between the primers was calculated. Function and analysis of the stability of the secondary structure of primer molecules, designed 6 sets of index primers containing Barcode, each set of 9 pairs of primers. Using these 6 sets of primers containing labels, the 6 DNA samples were amplified by LR-PCR respectively. For the three amplification reactions of fragments 1, 2, and 3, multi-tube amplification was used due to the low yield, and single-tube amplification was used for the others.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com