Screening method for anti-tumor medicine biomarker and application of anti-tumor medicine biomarker
A technology of anti-tumor drugs and biomarkers, applied in the field of tumor treatment, can solve the problems of non-recording algebra, noise, unclear algebra, etc., to improve efficiency and ability, reduce the impact of noise, and achieve the effect of small differences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0188] Example 1 Establishment of Liver Cancer Tumor Cell Line Collection and Chromosome Identification
[0189] After the solid tumor was isolated from multiple regions of the orthotopic tumor tissue of a liver cancer patient, a primary liver cancer cell line OPP containing 26 strains that can be stably passaged was successfully established after multiple times of defibrosis treatment and subculture. The cell name numbers are HCC307 2-1#, HCC307 2-2#, HCC307 2-3#, HCC307 2-4#, HCC307 2-5#, HCC307 5-1#, HCC307 5-3#, HCC307 7- 2#, HCC307 7-3#, HCC307 7-4#, HCC307 7-7#, HCC307 7-9#, HCC307 11-5#, HCC307 11-8#, HCC307 12-1#, HCC307 12-2# , HCC307 12-3#, HCC307 12-4#, HCC307 12-5#, HCC307 12-6#, CC307 12-7#, HCC307 13-1#, HCC307 13-4#, HCC307 13-6#, HCC307 14-1#, HCC307 14-2#.
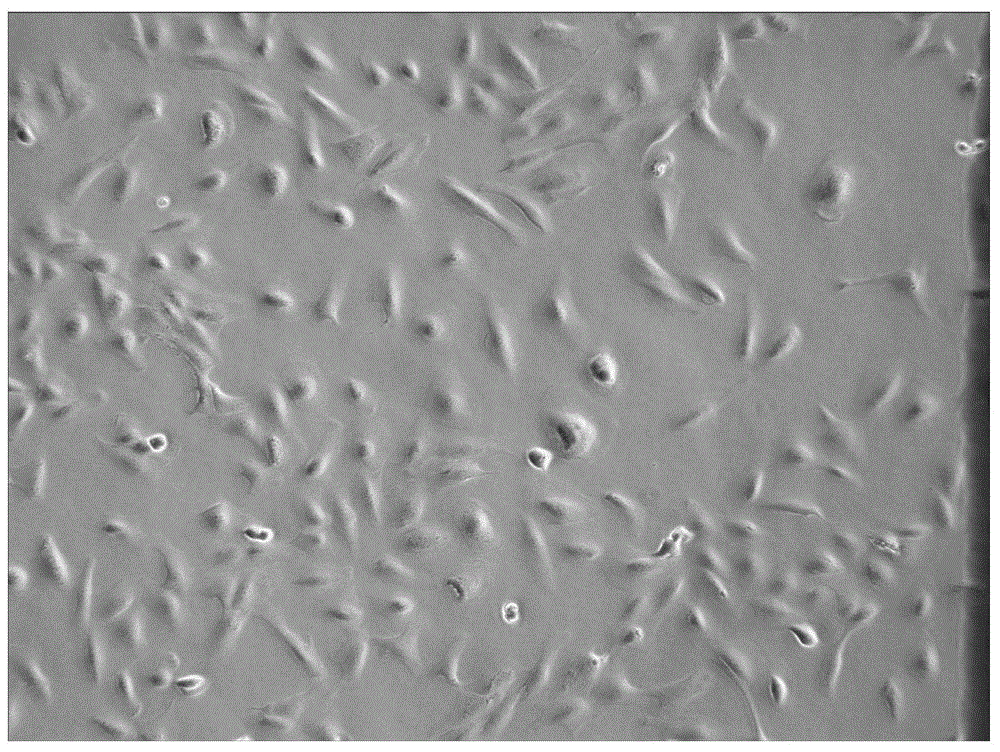
[0190] Observed from the cell morphology, the 26 cell lines all showed epithelial morphology (one representative cell line was as follows: figure 2 shown). After identification by karyotype analysis, ...
Embodiment 2
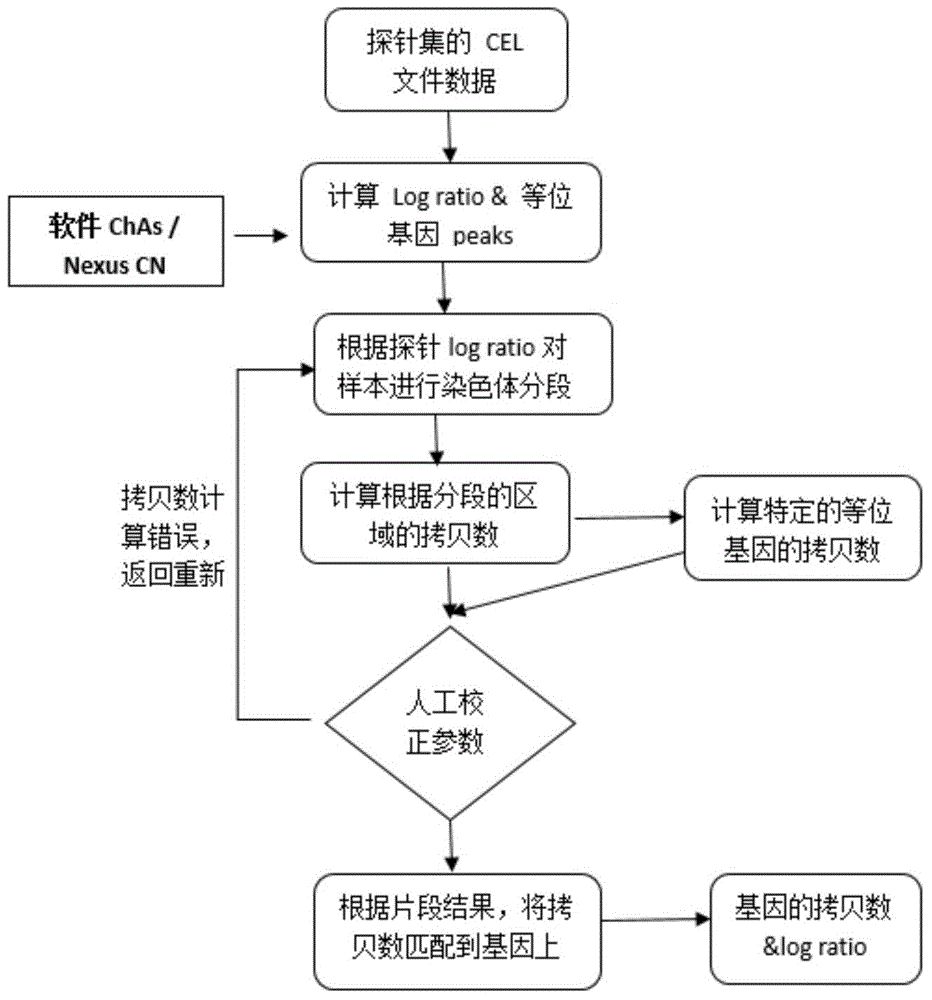
[0195] Example 2 Genome Sequencing
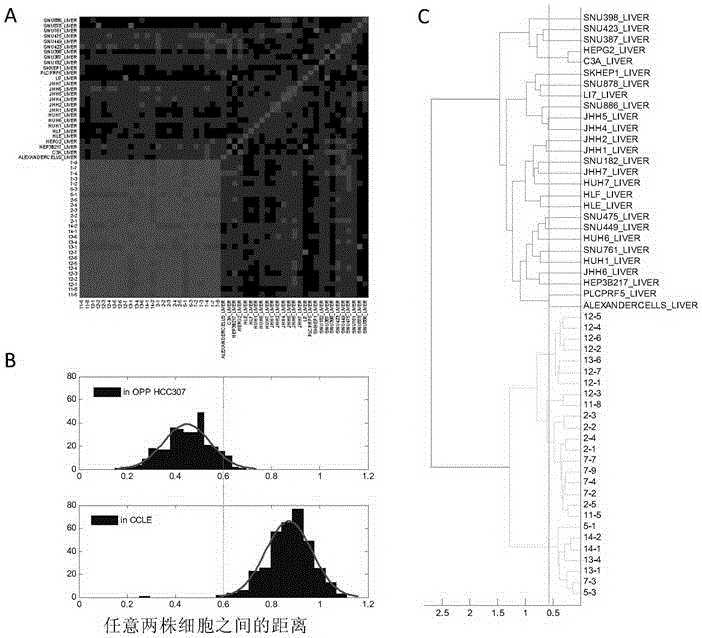
[0196] The gene expression profile, CNV copy number and exon sequencing of the 26 cell lines established in Example 1. The results of the data show that the genomes of these cell lines from the same subject have a high degree of homology, and there are also certain differences in gene expression, CNV copy number variation and exon variation. As shown in the figure, by comparing the CNV differences between cell lines, the CNV copy numbers among the 26 cell lines in OPP-HCC307 are not completely consistent, and there are differences, but the differences between them are significantly smaller than those from different subjects. Liver cancer cell lines (P=1.0953e-260, image 3 ).
[0197] image 3 It shows the evaluation of the distance between any two cell lines from different sources (including 26 OPP-HCC307 cell lines and 27 hepatoma cell lines from CCLE) using gene-based CNV data.
[0198] in image 3 A shows the heat map of (1-distanc...
Embodiment 3
[0199] Example 3 Antitumor Drug Sensitivity Test
[0200] Antitumor drug sensitivity tests were performed on the cell line collection obtained in Example 1. In the present embodiment, the antineoplastic drugs adopted are as follows:
[0201] Targeted drugs: 17-AAG, 2-deoxyglucose, Abiraterone, ABT-263, AC-220, AT-406, AZD4547, AZD5363, AZD7762, BI-2536, Birinapant, BMS-754807, Bortezomib (Bortezomib), BX-795, Cabozantinib, CAL-101, Carfilzomib, Crizotinib, Danusertib, Dasatinib, Dovitinib, Elesclomol, Embelin, Entinostat (MS-275), Enzastaurin, Everolimus, Foretinib, Fulvestrant, Ganetespib, GDC0941, GSK429286A, JQ1, KU-55933, KX2-391 , Lenvatinib, Linifanib, Linsitinib, LY2157299, Masitinib, MK-1775, MK-2206, MLN-4924, Neratinib, NU7441, Nutlin-3, NVP-BEZ235, NVP-BKM120, Oratinib, PCI-32765, PD-0325901, PD -0332991, PD-173074, PH-797804, PRT062607, R-406, Refametinib, Regorafenib, SCH900776, sgi-1776, Sorafenib, Sunitinib, TAE684 , Temsirolimus, TG-101348, Tideglusib, Tipi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com