Splicing method and system of second generation and third generation genomic sequencing data combination
A technology of genome sequencing and splicing system, which is applied in the splicing method and system field of second-generation and third-generation genome sequencing data, which can solve the difficulties of splicing prokaryotes and eukaryotes, reduce sequencing time and cost, and solve difficult repetitive sequences Area and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0047] The following are the concrete steps of the present invention, as follows:
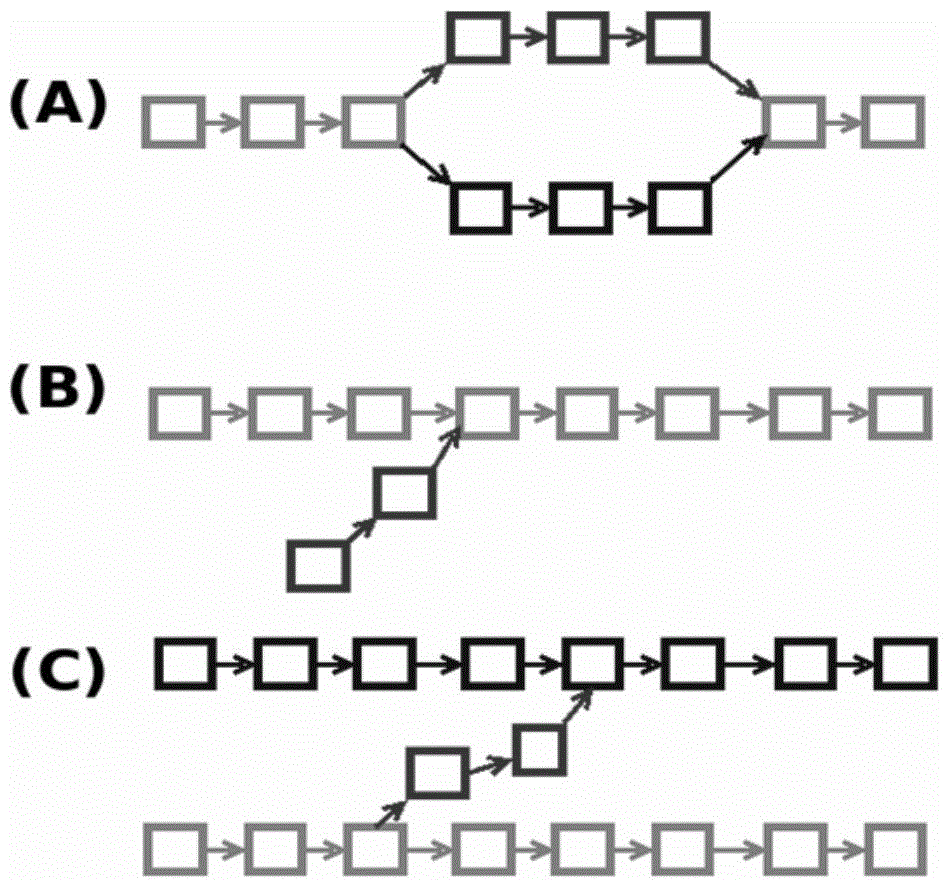
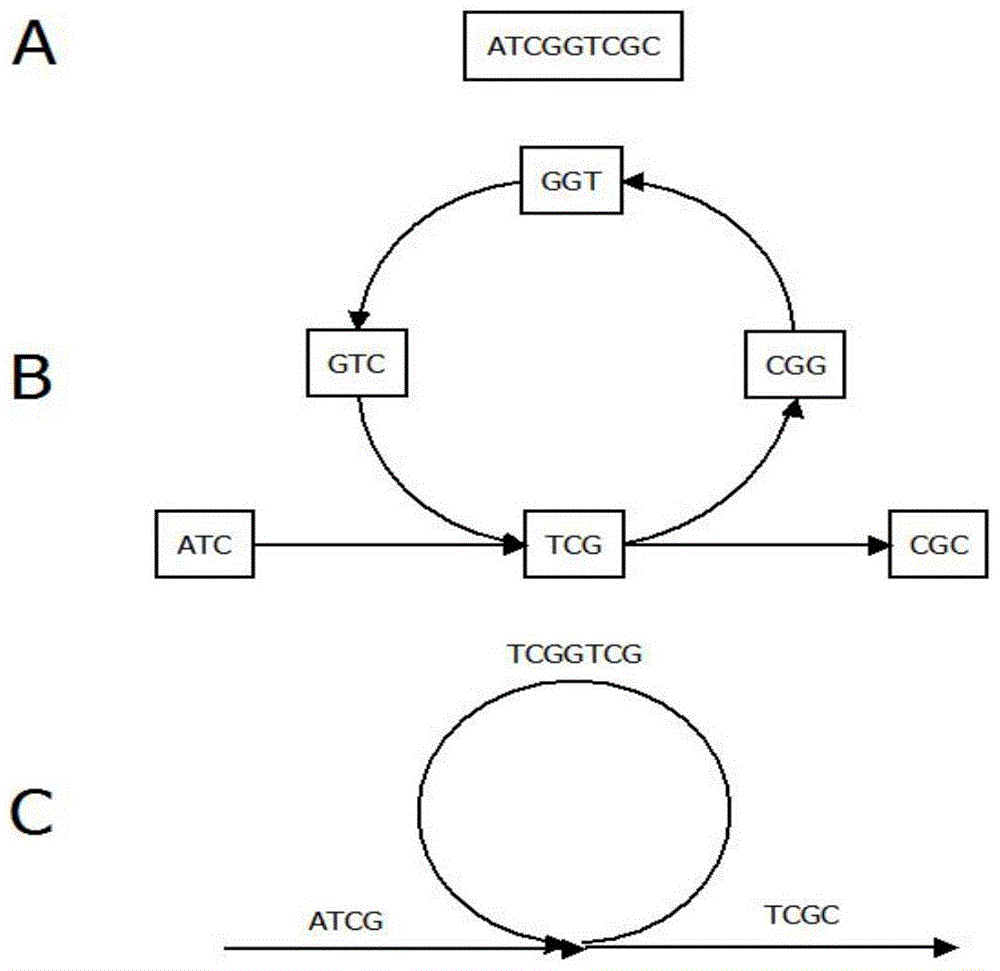
[0048] Step 1: Use the next-generation sequencing data to form a de Bruijn map. The second-generation sequencing data generally contains reads quality information. First, the quality information is used to preprocess the sequencing data to remove low-quality fragments, and then the reads are broken into kmers of the same length (k-mer refers to a reads, continuous Cutting, a sequence of nucleotide sequences with a length of K obtained by scratching one by one), constructing a de Bruijn graph, ARCS23 in the process of reading reads, generating a kmer according to the kmer length k input by the user, and saving it to the hash table , record the number of occurrences of kmer. In the implementation code of SOAPdenovo2, 1 byte is used to represent the number of occurrences of kmer. This method can only save up to 255. In the second-generation sequence splicing, the general sequencing depth will be r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com