Circular 'connection-extension' genome sequencing method
A genome sequencing and ligation technology, applied in the field of biomedicine, can solve the problems of high sequencing cost, difficulty in resequencing the human whole genome with high coverage, etc., and achieve the effect of great application prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0055] The present invention will be further described below in conjunction with the accompanying drawings and embodiments.
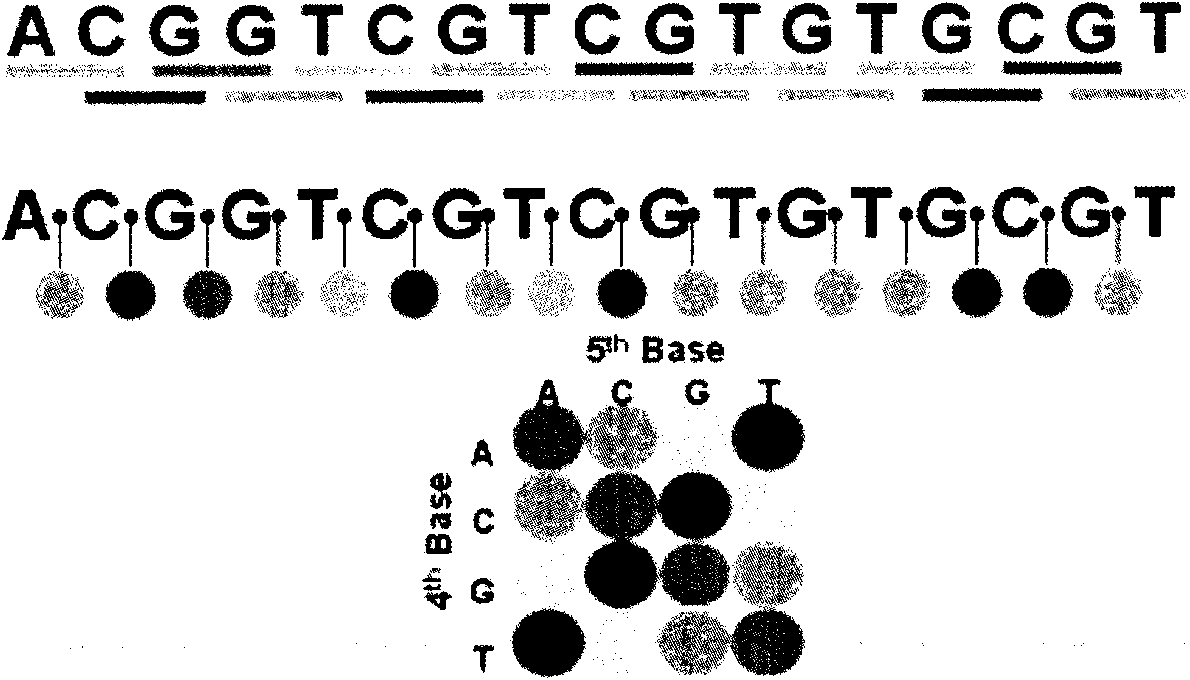
[0056] see figure 1 , the genome is composed of four bases: A, T, C, and G. Combining the four bases in pairs, we artificially define the following four-color definition matrix, so that we can combine all of a sequence Adjacent bases are defined by colors, and fluorescently labeled probes of corresponding colors are used for sequencing.
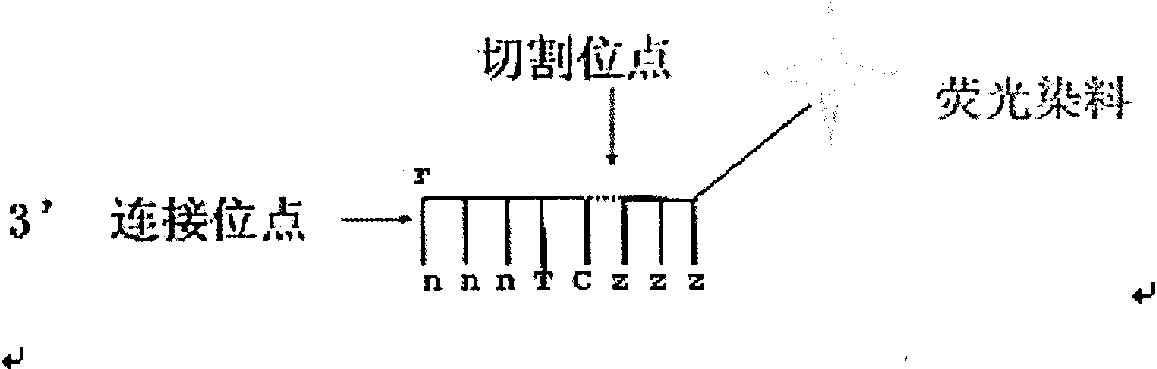
[0057] see figure 2 , the length of the specific probe is eight bases, and the fourth and fifth bases are specific binding sites. When the bases at these two positions are complementary to the bases at the corresponding positions of the template strand, A fluorophore attached to the probe tail then emits light.
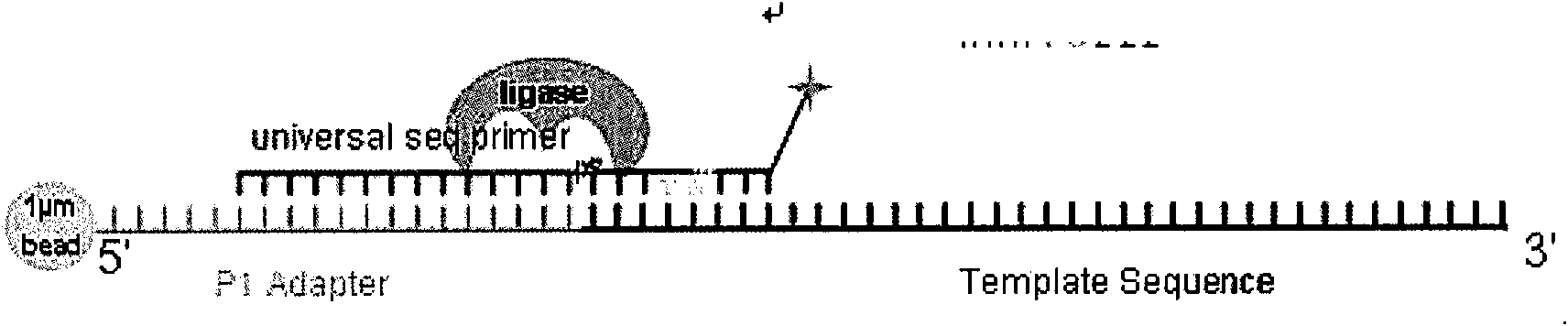
[0058] see image 3 , is the sequencing step (1) the universal primer is connected to the initial fragment of the template strand, the probe mixture reacts with the template, and the fourth and fifth pro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com