Long noncoding RNA, primer pair for detecting expression level of long noncoding RNA in cell line and tissue as well as kit
A long-chain non-coding, primer pair technology, used in DNA/RNA fragmentation, recombinant DNA technology, microbial determination/inspection, etc., can solve problems such as unclear functions, and achieve simple operation, high sensitivity, and good stability. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] 1. Template preparation (total RNA extraction and reverse transcription process)
[0031] Preparation of RNA: Colon cancer tissue and paired paracancerous tissue samples, gastric cancer tissue and paired paracancerous tissue samples were obtained. In this embodiment, 8 pairs of colon cancer tissues and paired paracancerous tissue samples are respectively CC1-CC8, CN1-CN8; and in this embodiment, 8 pairs of gastric cancer tissues and paracancerous tissue samples are respectively GC1-GC8, GN1 -GN8. The TRIZOL extraction method was used to extract the total RNA from the above samples, the absorbance value (A) was measured by an ultraviolet spectrophotometer to determine its content and purity, and the quality of the total RNA was identified by 0.8-1% agarose gel electrophoresis.
[0032] cDNA synthesis: 5 μg of the total RNA extracted from the above-mentioned colon cancer tissue and paired paracancerous tissues and gastric cancer tissues and paired paracancerous tissues w...
Embodiment 2
[0069] Example 2: Clinical Specimen Detection
[0070] (1) 20 pairs of colon cancer tissues and paired paracancerous tissues were taken, and Lnc5q21.2 was amplified by RT-PCR. Template preparation, PCR amplification system and conditions, and detection of amplified products were the same as those in Example 1. Lnc5q21. 2 Relative expression levels in colon cancer paired tissues, the test results are shown in Table 1 below:
[0071] Table 1
[0072]
[0073] (2) Take 20 pairs of gastric cancer tissues and paired paracancerous tissues, and use RT-PCR to amplify Lnc5q21.2. Template preparation, PCR amplification system and conditions, and detection of amplification products are the same as in Example 1, and Lnc5q21.2 is detected. The relative expression levels in gastric cancer paired tissues, the detection results are shown in Table 2 below:
[0074] Table 2
[0075]
Embodiment 3
[0076] Embodiment 3: Sensitivity experiment
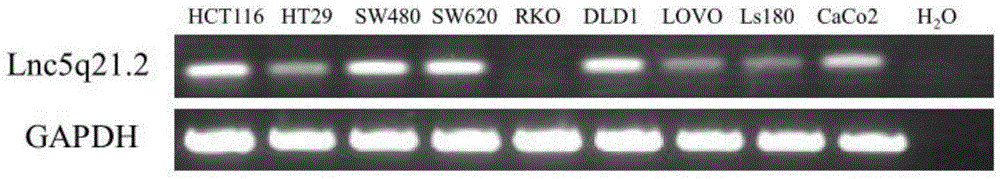
[0077] The cDNA samples prepared by the colon cancer cell line HCT116 with high expression of Lnc5q21.2 were selected and ddH 2 O was mixed in proportion, and the above-mentioned Lnc5q21.2 semi-quantitative RT-PCR primer pair was used to detect the expression level of Lnc5q21.2. Amplified products were subjected to agarose gel electrophoresis, such as Figure 5 shown.
[0078] The dilution ratios of the cDNA samples of the colon cancer cell line HCT116 were 100%, 50%, 5%, 1%, 0.5%, and 0%. wxya 2 O is the system control, used to evaluate whether there is contamination of PCR products in the system, such as ddH 2 If the result of O test is negative, the result of the system is credible, and the size of the amplification product of this system is 119bp.
[0079] Figure 5 The results show that: using the specific primer pair, reaction system and conditions for detecting the expression of Lnc5q21.2 in the present invention, the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com