Novel application of KCTD12 protein in cell cycle control
A cell cycle, new application of technology, applied in the field of protein science
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Effect of KCTD12 on cell cycle in S phase
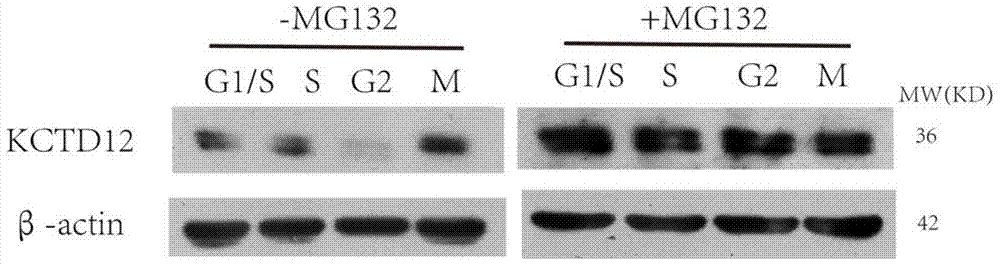
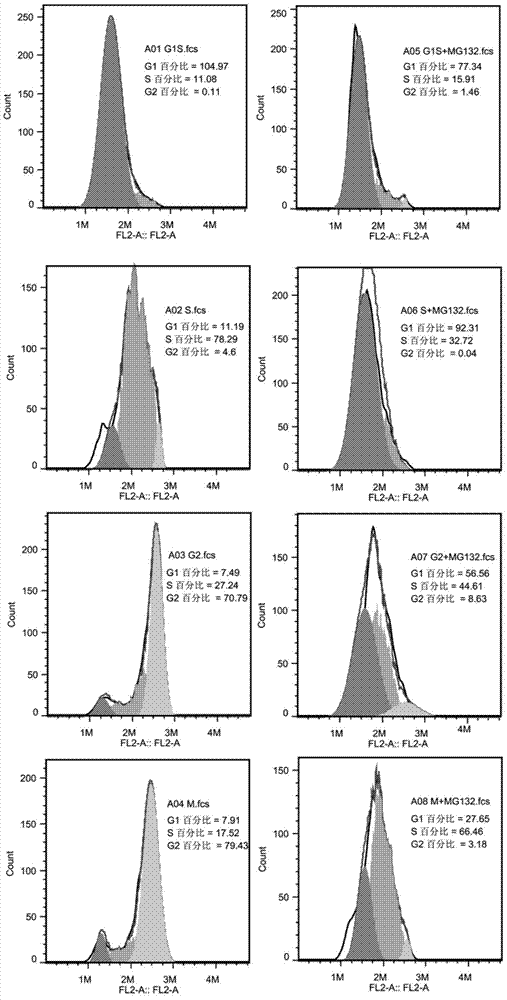
[0041] Synchronize the cells in the G1 / S phase, and add MG132 at a final concentration of 10 μM to treat the cells while releasing. MG132 is a proteasome inhibitor that inhibits proteasome degradation of ubiquitinated proteins. The cells of each phase were collected according to the method of synchronization, and the expression level of KCTD12 of the cells of each phase was detected by the method of Western Bolt ( figure 2 ). Then flow cytometry was used to detect the collected cells at each stage to detect the changes in their cycle ( image 3 ).
[0042] MG132 is a proteasome inhibitor that can inhibit the proteasome from degrading ubiquitinated proteins. figure 2 It indicated that MG132 significantly inhibited the degradation of KCTD12 in S phase and G2. image 3 It indicated that the addition of MG132 inhibited the degradation of protein KCTD12 and significantly blocked the process of cell cycle.
[0043]...
Embodiment 2
[0052] Example 2 Phosphorylation modification of KCTD12 under the regulation of Aurora kinase in M phase
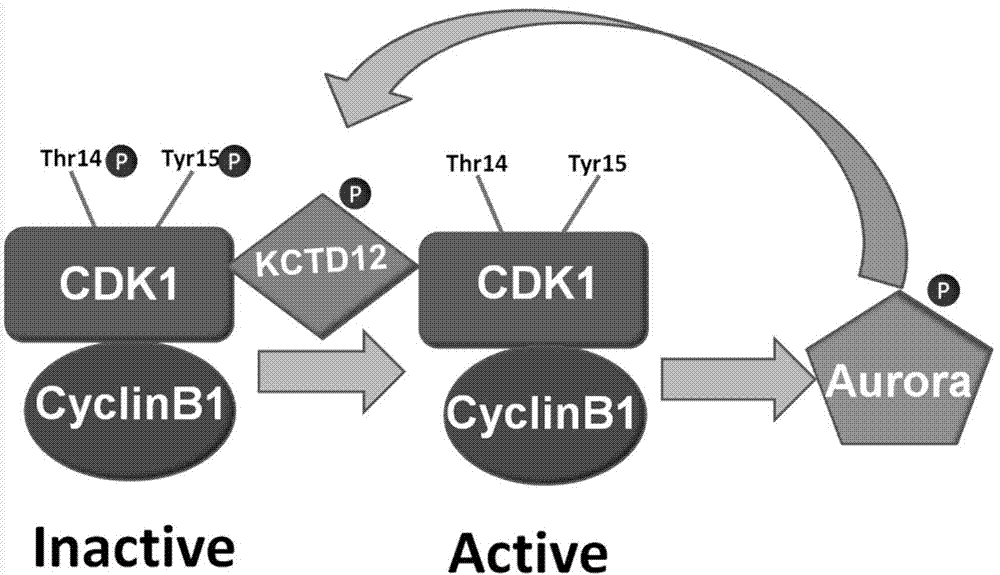
[0053] Since an important means of cell cycle regulation is to phosphorylate or dephosphorylate the protein to activate or inhibit the activity of the protein to achieve the purpose of regulation, it is speculated that KCTD12 may be phosphorylated, resulting in a larger molecular weight , the position moves up (in Western Bolt). In order to confirm the speculation, the protein in the M phase was treated with alkaline phosphatase (CIP), and the results Figure 4 It was shown that the position of KCTD12 shifted down after CIP treatment. It was confirmed that KCTD12 shifted up in M phase because of phosphorylation modification.
[0054] After confirming that KCTD12 undergoes phosphorylation modification in the M phase, it is found that KCTD12 has an Aurora kinase action site by searching the database of phosphorylated proteins and comparing the motifs. From this, it w...
Embodiment 3
[0055] Example 3 Serine at position 243 is the phosphorylation site of KCTD12
[0056] The results of Example 2 confirm that KCTD12 is regulated by Aurora kinase in the M phase and undergoes phosphorylation modification. According to the pre-stored phosphorylation site, construct its mutation vector to determine its phosphorylation site. First construct the recombinant plasmid of pCMV-N-flag-KCTD12 on the backbone of pCMV-N-flag, and then perform point mutation on the basis of the already constructed pCMV-N-flag-KCTD12 to construct the phosphorylation site Mutation vector (mut1).
[0057] The plasmid pCMV-N-flag was used as the backbone to construct pCMV-N-flag-KCTD12. KCTD12 uses the cDNA of HeLa cells as a template, and designs primers for PCR amplification. The length of KCTD12 gene PCR product is about 1kb. Both the PCR product and the plasmid were digested with EcoR I and XbaI at 37°C for 4 hours, ligated overnight at 16°C, then transformed with E.coli competent, plate...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com