Method for detecting chromosome number variation of fetus
A technology of chromosome number and fetus, applied in the field of chromosome detection, can solve the problems of PCR amplification bias, uneven GC content of the genome, affecting the PCR amplification efficiency, etc., so as to reduce the investment of reagents, primers and time, and save the Effects of PCR amplification procedures, simplified library construction process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Example 1 Method for constructing sequencing library
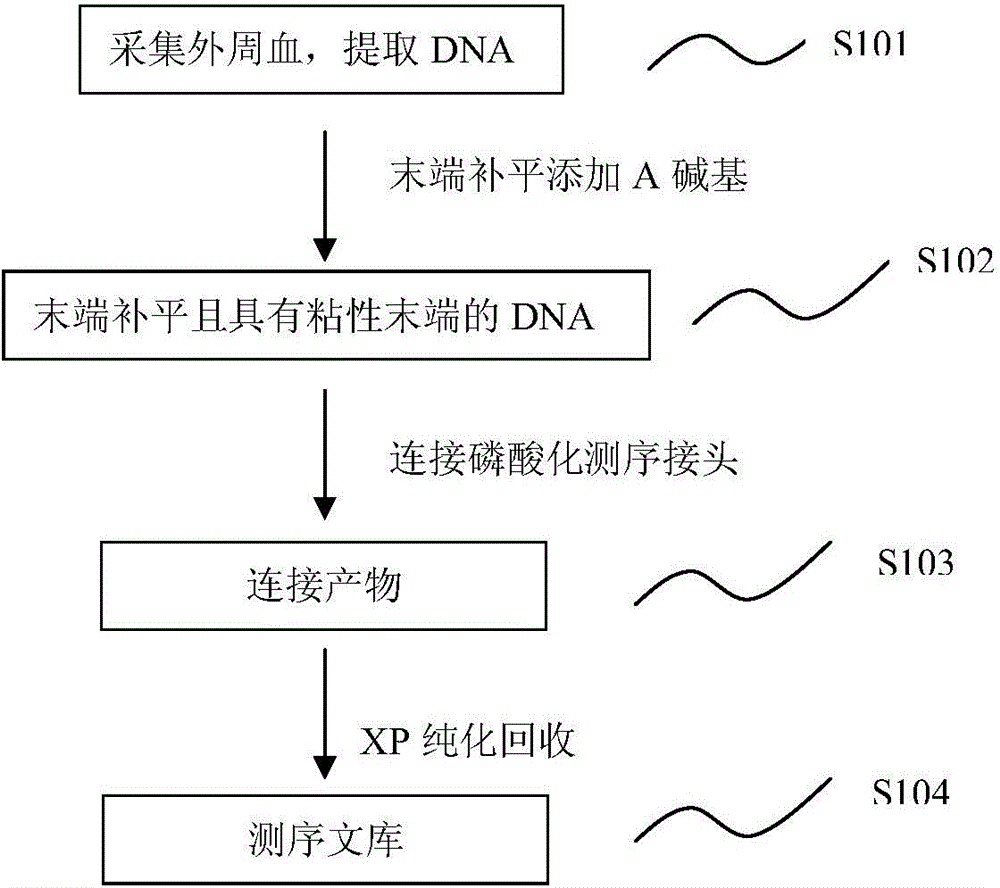
[0063] According to one aspect of the present invention, the present invention provides a method for constructing a sequencing library, such as figure 1 As shown, according to an embodiment of the present invention, the method includes the following steps:
[0064] Step S101, extract peripheral blood to obtain DNA in plasma. The term "DNA" used in the present invention can be any polymer comprising any deoxyribonucleotides, including but not limited to modified or unmodified DNA. Those skilled in the art can understand that the source of DNA is not particularly limited, and can be obtained from any possible way, it can be obtained directly from the market, it can also be obtained directly from other laboratories, and it can also be obtained directly from samples. extract from. According to an embodiment of the present invention, genomic DNA can be extracted from a sample. According to an embodiment of the presen...
Embodiment 2
[0069] Example 2 kit
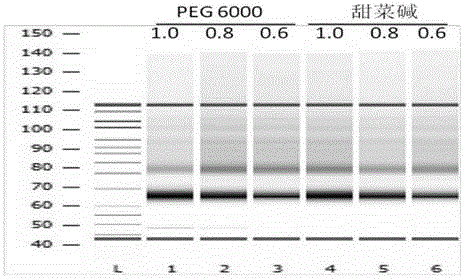
[0070] According to another aspect of the present invention, the present invention provides a sequencing library kit for constructing a specific region of the genome of a sample. According to an embodiment of the present invention, the kit may include an enzyme for DNA end repair plus A base And the required buffer containing Tris-HCl, KCl, MgCl2, DDT, dNTPs; the ligase used to connect the modified DNA and the sequencing adapter and the required buffer containing Tris-HCl, MgCl2, DTT, ATP, betaine Buffers, connectors. Those skilled in the art can understand that the kit may further include any other components required for constructing the sample sequencing library.
[0071]The method for constructing a sequencing library of the present invention omits the step of PCR amplification, so the method for constructing a sequencing library of the present invention reduces the deviation generated by PCR amplification and improves the accuracy of sequencing. Be...
Embodiment 3
[0073] Example 3 The specific operation process of constructing the sequencing library
[0074] 1. Preparation of plasma DNA
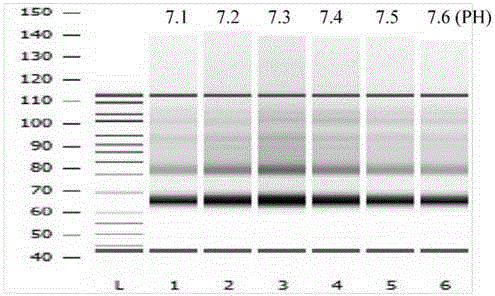
[0075] The peripheral blood of 10 pregnant women was drawn, and the numbers were: 14N0001, 14N0002, 14N0003, 14N0004, 14N0005, 14N0006, 14N0007, 14N0008, 14N0009 and 14N0010. Blood samples were centrifuged in two steps to obtain plasma without blood cells, and sterilized high-purity water was used instead of plasma as a negative control, and the gold standard amniocentesis confirmed sample was used as a positive control, and the volume of each sample was 1ml.
[0076] Use the serum / plasma free DNA extraction kit produced by TIANGEN to extract DNA in plasma (QIAamp circulating nucleic acid kit from Qiagen or other companies’ DNA extraction kits are also applicable to the present invention), and the operation process is carried out according to the instructions.
[0077] 2. Repair of plasma DNA ends and addition of base A
[0078] The extracted DNA is ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com